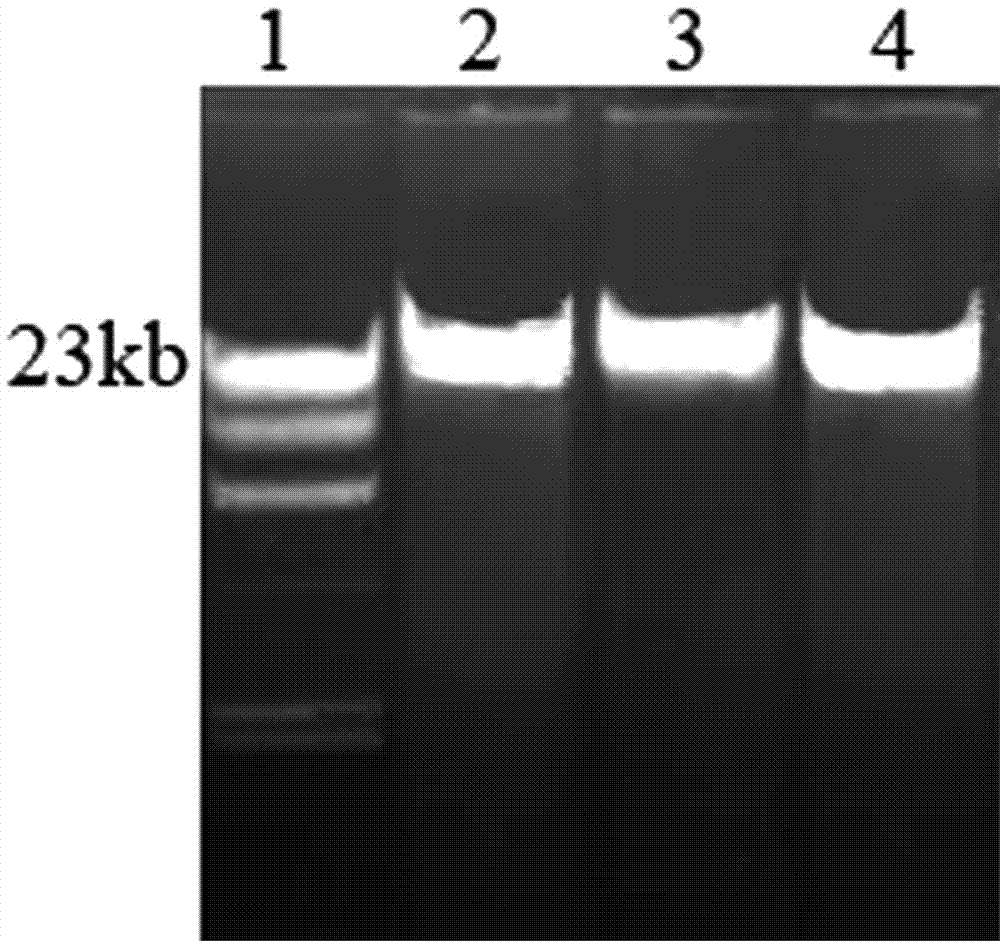
Method for rapidly extracting microbial genomes from soil sample
A technology for soil microorganisms and soil samples, applied in the field of rapid extraction of microbial genomes in soil samples, can solve the problems of analyzing the impact of biological analysis, and achieve the effects of high DNA integrity, time saving, and simple reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Soil sample: the soil around the wheat root system in the wheat field, when the wheat was mature and the sickle was about to open.
[0032] The specific process of separating and extracting the soil microbial genome is as follows:
[0033] (1) Grind and pulverize 2g of soil sample in liquid nitrogen;
[0034] (2) Put the ground sample in a 10ml centrifuge tube, add genome extraction buffer (including 0.2MNaCl, 0.05MEDTA, 0.15Tris-HCl and 1mg / ml lysozyme), shake for 3min, mix well, and place in a 37°C water bath Incubate in medium for 30min;
[0035] (3) Add proteinase K solution and SDS solution to the solution to a final concentration of 0.02 mg / ml and 0.2%, mix slightly, and bathe in 55°C water for 30 minutes;
[0036] (4) Centrifuge at 10,000 rpm for 5 minutes at 4°C, and collect the supernatant;
[0037] (5) The supernatant was extracted with an equal volume of phenol at room temperature for 5 minutes;
[0038] (6) Centrifuge at 10,000 rpm for 5 minutes at 4°C, ...
Embodiment 2
[0046] Soil sample: the soil around the corn root system in a cornfield, and the sampling time is the jointing stage of corn growth.
[0047] The specific process of separating and extracting the soil microbial genome is as follows:
[0048] (1) Grind and pulverize 5g of soil sample in liquid nitrogen;
[0049] (2) Put part of the ground sample in a 10ml centrifuge tube, add genome extraction buffer (including 0.2MNaCl, 0.05MEDTA, 0.15Tris-HCl and 2mg / ml lysozyme), shake for 3min, mix well, and store at 37°C Incubate in a water bath for 30 minutes;
[0050](3) Add proteinase K solution and SDS solution to the solution to a final concentration of 0.03 mg / ml and 0.3%, mix slightly, and bathe in 55°C water for 30 minutes;
[0051] (4) Centrifuge at 10,000 rpm for 10 minutes at 4°C, and collect the supernatant;
[0052] (5) The supernatant was extracted with an equal volume of phenol at room temperature for 5 minutes;
[0053] (6) Centrifuge at 10,000 rpm for 10 minutes at 4°C...
Embodiment 3
[0061] Soil sample: the soil around the tomato root system in the tomato planting greenhouse, and the sampling time is the flowering and fruit-setting period of tomato growth.
[0062] The specific process of separating and extracting the soil microbial genome is as follows:
[0063] (1) Grind and pulverize 1g of soil sample in liquid nitrogen;
[0064] (2) Put part of the ground sample in a 10ml centrifuge tube, add genome extraction buffer (including 0.2MNaCl, 0.05MEDTA, 0.15Tris-HCl and 2mg / ml lysozyme), shake for 3min, mix well, and store at 37°C Incubate in a water bath for 30 minutes;
[0065] (3) Add proteinase K solution and SDS solution to the solution to a final concentration of 0.05mg / ml and 0.5%, mix slightly, and bathe in 55°C water for 30 minutes;
[0066] (4) Centrifuge at 12,000 rpm for 10 minutes at 4°C, and collect the supernatant;
[0067] (5) The supernatant was extracted with an equal volume of phenol at room temperature for 5 minutes;
[0068] (6) Cen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com