A method for expressing biotinylated luciferase
An expression method, luciferase technology, which is applied in the field of expression of biotinylated DNA polymerase, can solve the problems of low ATPS activity of fusion protein, reduce the expression level of fusion protein, and cannot better meet the practical application, so as to increase the yield , the effect of activity improvement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Embodiment 1 Expression method of biotinylated luciferase
[0036] Include the following steps:
[0037] ⑴. Select the North American firefly luciferase whose GenBank number is CAA46407, and design a pair of specific primers: the nucleotide sequence of the forward primer is shown in SEQ ID NO: 3, and the restriction site is BamHI; The nucleotide sequence is shown in SEQ ID NO: 4, and the enzyme cutting site is XhoI;
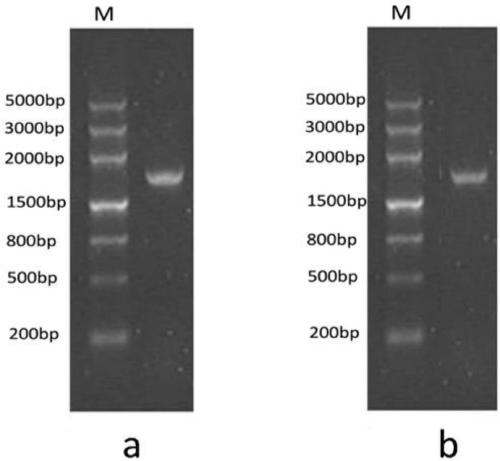
[0038] (2) The forward primer and reverse primer described in step (1) of chemical synthesis embodiment 1, with - luc vector as a template, amplify the fusion coding gene of the biotin acceptor peptide of the present invention and the luciferase described in step (1) of Example 1 by PCR, the PCR program is: denaturation at 95°C for 10 minutes, denaturation at 95°C for 30s , anneal at 50°C for 30s, extend at 72°C for 1.5min, 35 cycles, and finally extend at 72°C for 10min;
[0039] ⑶. The DNA gel recovery kit reclaims the fusion coding gene obtained in ...
Embodiment 2
[0042] Embodiment 2 Purification of biotinylated luciferase
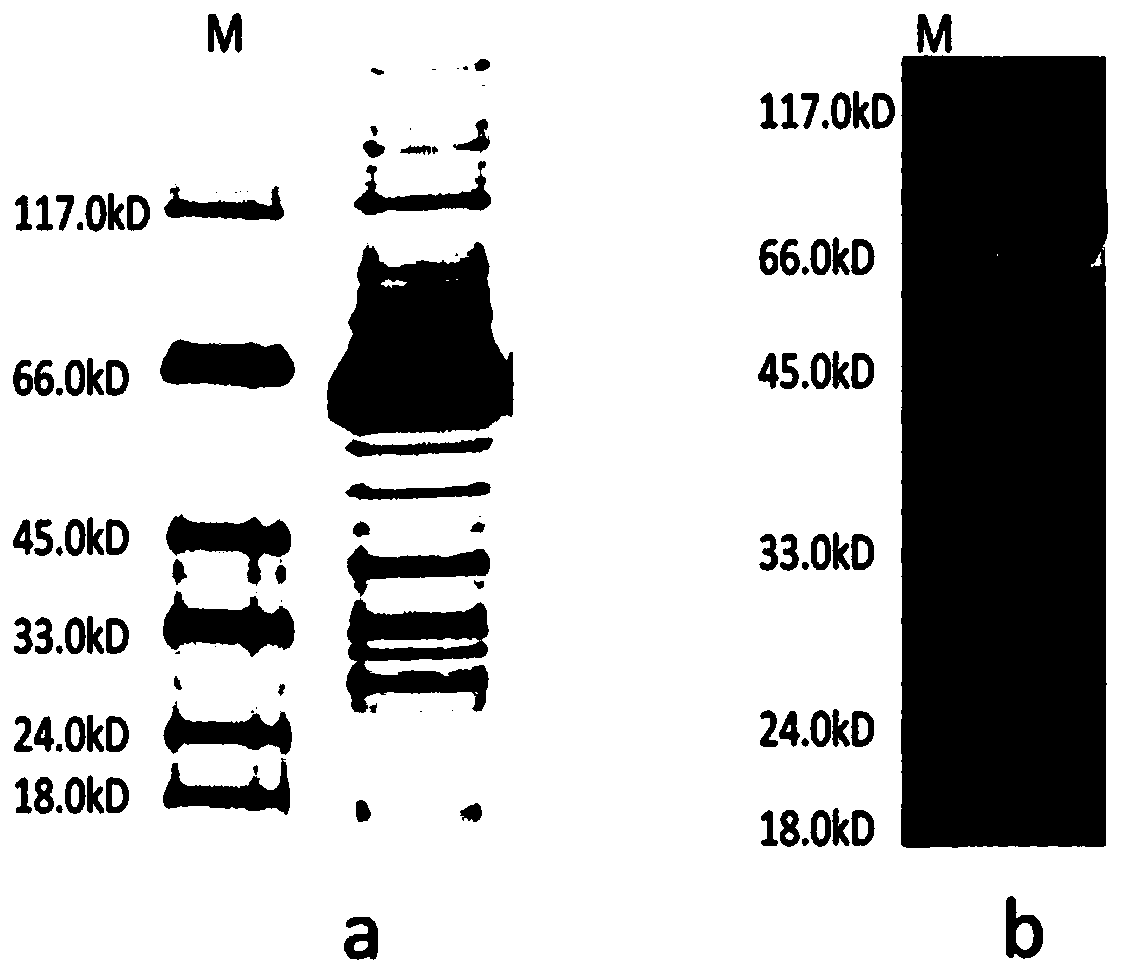
[0043] The biotinylated luciferase expressed according to the method of Example 1 was purified using a histidine affinity chromatography column.
[0044] The purification method comprises the following steps:
[0045] ⑴. Centrifuge to collect the bacterial cells after the induction culture in step ⑸ of Example 1, crush the bacterial cells by ultrasonic waves, centrifuge the crushed liquid at 4°C and 12000rpm / min for 30min, and collect the supernatant;
[0046] ⑵. Equilibrate the histidine affinity chromatography column with 5 times the volume of the column bed (50mM pH8.0 sodium phosphate buffer + 300mM sodium chloride + 5mM imidazole);
[0047] (3) The supernatant obtained in step (1) of Example 2 was added to a histidine affinity chromatography column at a flow rate of 1 mL / min, so that biotinylated luciferase was attached to the column;
[0048] ⑷. Rinse the chromatography column with 15 times the volume of the...
Embodiment 3
[0051] Embodiment 3 Expression method of biotinylated luciferase
[0052] Include the following steps:
[0053] ⑴. Select a heat-resistant mutant rt-LlL of Japanese firefly luciferase, which is replaced by leucine by replacing 217 alanine (Ala) in Japanese firefly luciferase with GenBank numbering X66919.1 acid (Leu) acquisition (Xu Shu, Zou Bingjie et al. Expression of thermostable luciferase and establishment of thermostable pyrosequencing system[J]. Acta Biological Engineering, 2012, 06:763-771); design a pair of specific Sexual primer: the nucleotide sequence of the forward primer is shown in SEQ ID NO: 5, and the restriction site is BamHI; the nucleotide sequence of the reverse primer is shown in SEQ ID NO: 6, and the restriction site is HindⅢ ;
[0054] (2) The forward primer and the reverse primer described in Step (1) of Example 3 were chemically synthesized, and the rt-L1L recombinant vector containing the coding gene of the heat-resistant mutant rt-L1L described in...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com