A kind of molecular detection primer and rapid detection method of maize leaf spot bacterium
A technology for the detection of maize leaf spot bacteria and molecules, which is applied in the field of crop disease detection and biology, can solve the problems of long-term consumption, high requirements for identification experience, and low accuracy, and achieve high sensitivity, wide applicability, and good practicability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Example 1 Molecular Detection Primer Design and Establishment of Specific Molecular Detection Method for Septoria maize
[0029] 1. Extraction of Genomic DNA of Pseudomonas maize:
[0030] The CTAB method was used to extract the genomic DNA of 6 strains of P. maize spp. preserved in our laboratory, and the specific steps were as follows:
[0031] (1) Take 0.1 g of mycelium powder in a 1.5 mL centrifuge tube, add 900 μL of 2% CTAB extract, shake and mix with a shaker, bathe in 60°C water for 60 min, and centrifuge at 12,000 r / min for 15 min at room temperature;
[0032] (2) Take 700 μL of the supernatant, add an equal volume of phenol, chloroform, and isoamyl alcohol mixture (the volume ratio of each is 25:24:1), shake gently, and centrifuge at 8000 r / min for 10 minutes at room temperature;
[0033] (3) Take 500 μL of the supernatant, add an equal volume of chloroform and extract again, and centrifuge at 8000 r / min for 10 min at room temperature;
[0034](4) Take 350 μ...
Embodiment 2
[0051] Example 2 Specific Amplification of Pseudomonas maize
[0052] 1. Using the CTAB method to extract 2 strains of different sources of maize leaf spot, maize gray spot, maize round spot, maize small spot, maize southern rust, maize sheath blight, rice blast fungus and peanut brown spot Genomic DNA of bacteria.
[0053] 2. Use the DNA extracted from the test bacteria as a template for PCR amplification: 25 μL of PCR reaction system, including 2.5 μL of 10×PCR buffer, 2.0 μL of dNTP Mixture with a concentration of 2.5 mmol / L, and 0.15 μL of Taq with a concentration of 5 U / μL Enzyme, 0.5 μL each of 10 μmol / L primers ESF and ESR, 1 μL DNA template, add ddH 2 O to a total volume of 25 μL; PCR reaction conditions: 94°C pre-denaturation for 5 min; 94°C denaturation for 45 sec, 55°C annealing for 45 sec, 72°C extension for 45 sec, a total of 35 cycles; 72°C extension for 10 min; electrophoresis detection of amplified products.
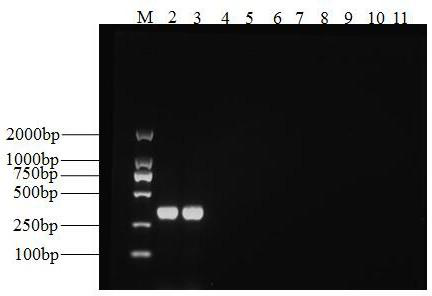
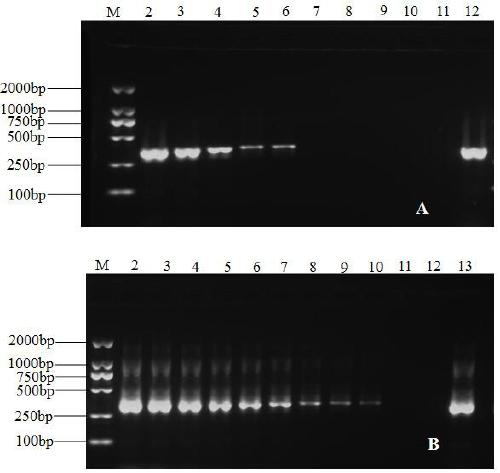
[0054] 3. Specific amplification results
[0055...
Embodiment 3
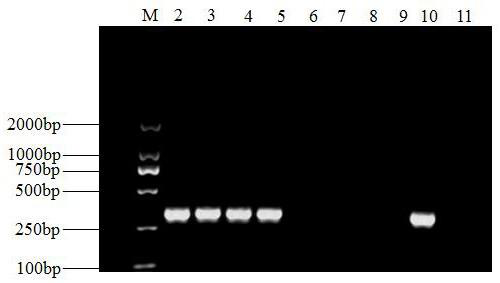
[0056] Example 3 Sensitivity detection of primers of the present invention to Pseudomonas maize
[0057] 1. Using the CTAB method to extract the genomic DNA of P. maize;
[0058] 2. After measuring the concentration of the extracted genomic DNA of Leptosphaeria maize with a spectrophotometer, dilute it with sterile ultrapure water to prepare a series of concentrations for subsequent use;
[0059] 3. Carry out routine PCR amplification using the prepared series of DNA as a template: 25 μL of PCR reaction system, including 2.5 μL of 10×PCR buffer, 2.0 μL of dNTP Mixture with a concentration of 2.5 mmol / L, and 0.15 μL of dNTP Mixture with a concentration of 5 U / μL Taq enzyme, 0.5 μL each of 10 μmol / L primers ESF and ESR, 1 μL DNA template, add ddH 2 O to a total volume of 25 μL; PCR reaction conditions: 94°C pre-denaturation for 5 min; 94°C denaturation for 45 sec, 55°C annealing for 45 sec, 72°C extension for 45 sec, a total of 35 cycles; 72°C extension for 10 min; electrophore...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com