Composite lactobacillus nipple cleaning solution for improving dairy cow nipple micro-ecosystem
A technology of compound lactic acid bacteria and micro-ecological system, applied in the field of micro-ecological preparations, can solve the problems of excessive iodine content in milk and poor effect of bovine mastitis bacteria, and achieve the effects of controlling the number of somatic cells, increasing milk production, and reducing production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0037] The preparation method of compound lactic acid bacteria nipple cleaning liquid of the present invention comprises the steps:
[0038] a. Activation of strains: Lactobacillus plantarum L.plantarumSCI-01 and L.plantarumSCI-02 frozen at -80°C were inoculated in MRS liquid medium and cultured at 37°C for 24 hours, and subcultured twice to obtain activated bacteria kind;
[0039] b. Preparation of fermentation medium: sucrose 50kg, yeast powder 20kg, soybean peptone 10kg, Tween-800.8kg, water 1000L, fully dissolved and then sterilized at 121°C for 15min;
[0040] c. Preparation of seed culture solution: inoculate the above-mentioned activated strains in the fermentation medium with an inoculum of 1% respectively, and carry out mixed fermentation culture at 37 ° C. During the fermentation process, the pH is controlled by adding ammonia water until the acid production stops. Terminate the fermentation when adding ammonia water again, and aseptically fill the fermented liquid ...
Embodiment 1
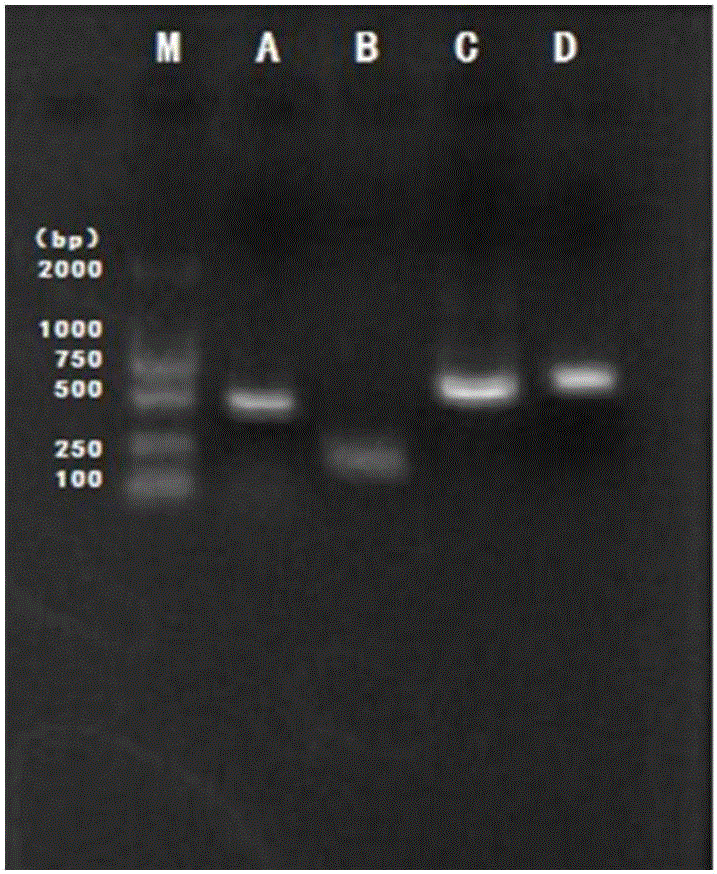
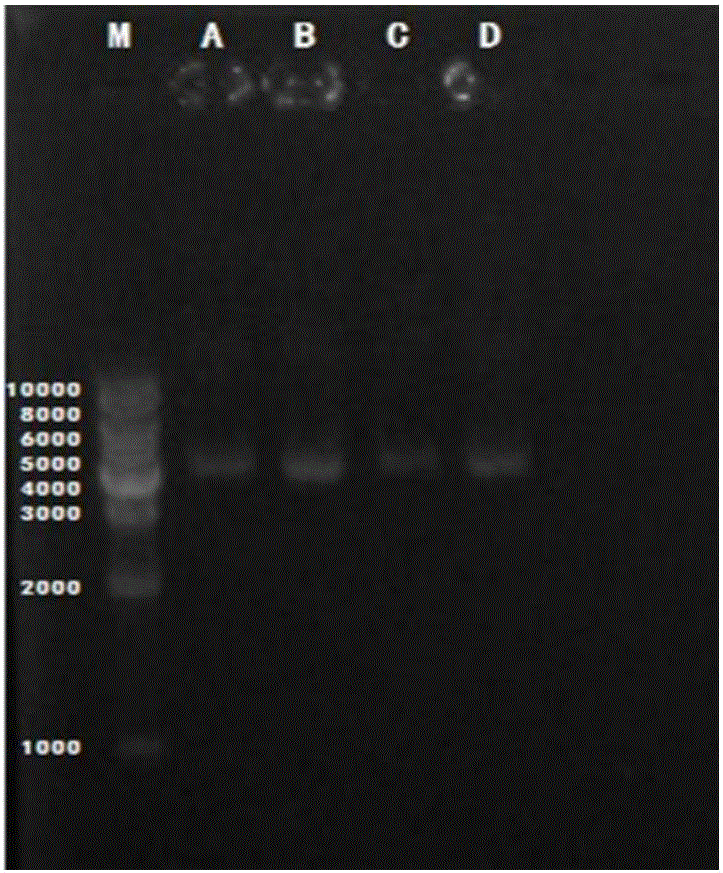
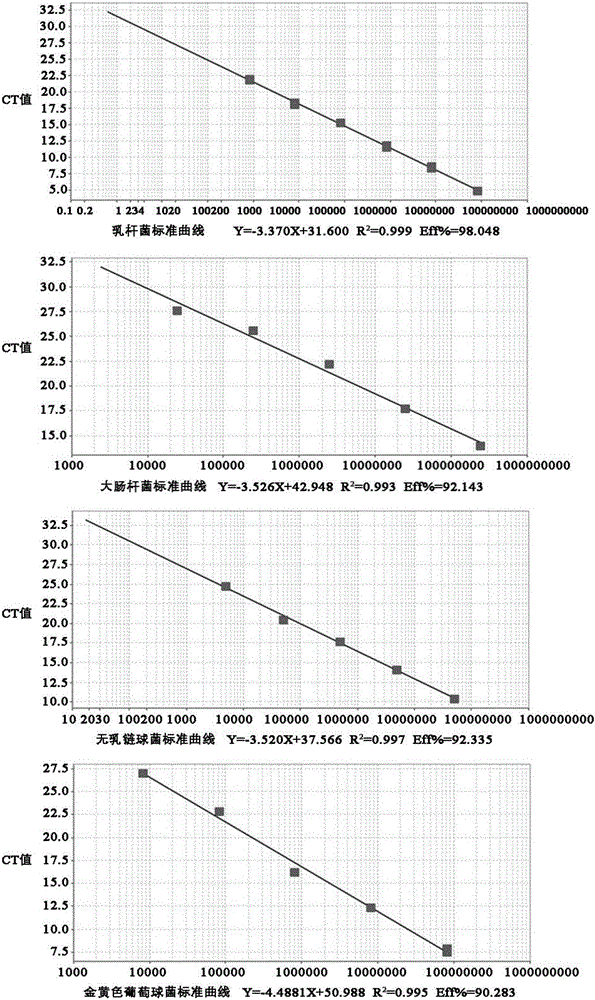
[0041] Embodiment 1 Quantitative detection of Lactobacillus, Escherichia coli, Streptococcus agalactiae and Staphylococcus aureus in milk samples by fluorescent quantitative PCR technology, real-time monitoring of the above-mentioned microorganisms in the trial process of the composite lactic acid bacteria teat cleaning solution of the present invention Changes in the content of milk. The Fourier online 400 system was used to measure the change of the number of somatic cells in milk during the trial process of the cleaning solution. The plate count method was used to detect the changes in the total number of colonies, lactic acid bacteria, coliforms, staphylococcus, shigella and salmonella in milk before and after the test.
Embodiment 2
[0042] Example 2 Fourier online 400 system was used to measure the changes of somatic cell number, lactose rate, protein rate and total solids in milk during the trial process of the cleaning solution of the present invention. The melanin test method was used to determine the total number of bacteria in milk according to the time of melanin fading. The changes in the daily milk production of the experimental cows before and after the statistical test were calculated.
[0043] research method:
[0044] 1. Extraction of metagenomic DNA from milk samples
[0045] Microbial metagenomic DNA in milk samples was extracted using the QIAampDNAStoolMiniKit kit. For specific methods, refer to the kit instruction manual. The concentration and quality of metagenomic DNA were detected by micro-volume ultraviolet spectrophotometer.
[0046] 2. Specific primer design
[0047] Four sets of specific primers were designed and synthesized for Lactobacillus, Escherichia coli, Staphylococcus au...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com