A kit for detecting hepatitis C virus nucleic acid using one-step real-time fluorescent quantitative RT-qPCR
A real-time fluorescence quantitative, hepatitis C virus technology, applied in the field of biomedical detection, can solve problems such as degradation, provide accurate evidence, and reduce detection efficiency, so as to reduce the probability of enzymatic degradation, eliminate false negative results, and ensure authenticity and reliability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Embodiment 1 Preparation of the kit for detecting hepatitis C virus nucleic acid according to the present invention
[0045] The kit for detecting hepatitis C virus nucleic acid by one-step real-time fluorescent quantitative RT-qPCR according to the present invention includes virus nucleic acid extraction reagents, probes for detecting target polynucleotides and internal controls and for amplifying target polynucleotides Primer RT-qPCR Reaction Buffer with Hot Start DNA Polymerase, Revert Aid TM M-MLV Reverse Transcriptase and RiboLock TM RT-PCR enzyme mix of ribonuclease inhibitors, quantitative standard reference containing pseudovirions,
[0046] Its preparation method is as follows:
[0047] 1. Preparation of Viral Nucleic Acid Extraction Reagent
[0048] a. RNA extraction solution I: guanidine isothiocyanate 40-60 g / 100 ml, disodium edetate 0.74-1.49 g / L, urea 0.6-1.5 g / L, Triton X-100 3- 5 ml / 100 ml, Tween-20 4-6 ml / 100 ml, after weighing the above-mentioned ...
Embodiment 2
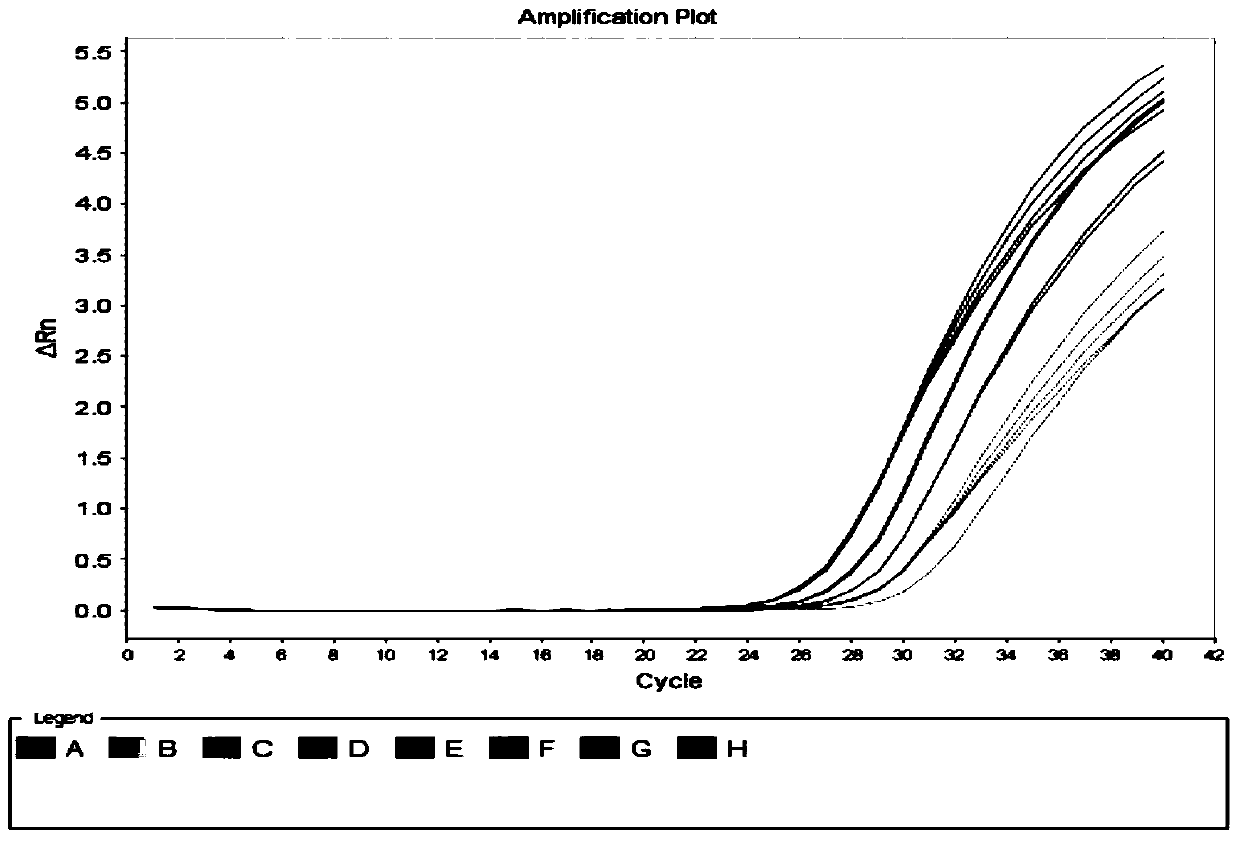
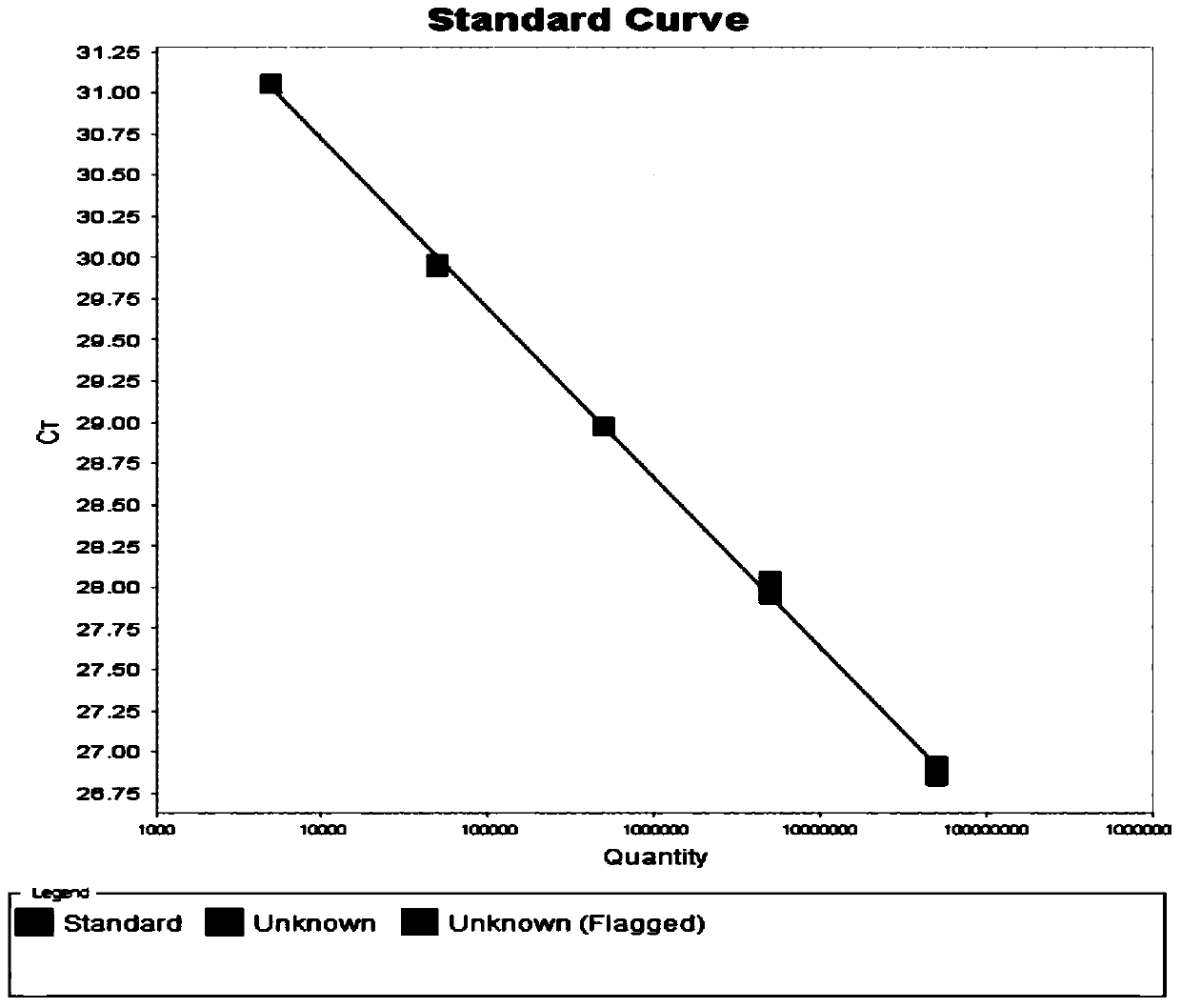
[0090] Embodiment 2 utilizes the hepatitis C virus nucleic acid quantitative detection kit of the present invention to detect the HCV-RNA concentration in the clinical serum sample
[0091] All positive samples in this experiment were clinical serum samples collected from Shandong Provincial Hospital, Shandong Qianfoshan Hospital and Shandong University Qilu Hospital. The HCV genotyping of the samples was performed by Daan Genotype Hepatitis C Virus (HCV) genotyping Detection kit (PCR-fluorescent probe method). See the table below for sample information:
[0092]
[0093]
[0094] Negative samples in this experiment include: normal human serum, hepatitis A virus antibody positive serum (anti-HAV + ), hepatitis B surface antigen positive serum (HBsAg + ), hepatitis E virus antibody positive serum (anti-HEV + ), human immunodeficiency virus antibody positive serum (anti-HIV + )sample. The specific information of each sample is shown in the table below:
[0095]
...
Embodiment 3
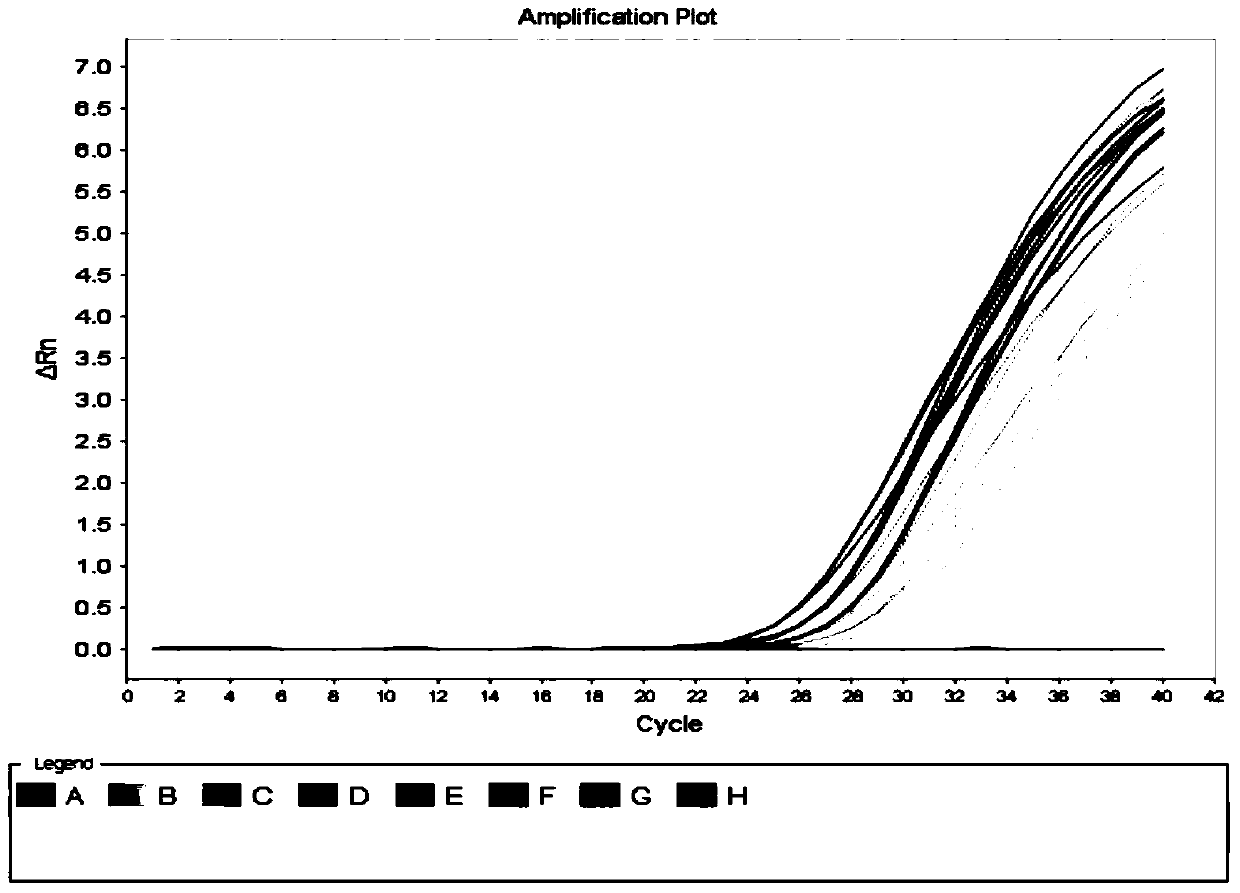
[0135] Example 3 Utilize the kit of the present invention to detect HCV-RNA concentration in 20 cases of hepatitis C nucleic acid positive serum samples before and after degradation
[0136] 1. Sample Processing
[0137] A. Processing of Stable HCV RNA Samples
[0138] The blood of the HCV-positive patients was centrifuged immediately after collection, and the serum was collected and divided into two parts. One copy was immediately frozen at -80°C as a stable HCV RNA sample.
[0139] B. Processing of unstable HCV RNA samples
[0140] The other serum above was placed at room temperature overnight, and then placed in a refrigerator at 4°C for 6 hours to simulate the irregular operation from sample collection to storage, as an unstable degraded HCV RNA sample.
[0141] 2. Extraction of sample nucleic acid
[0142] 1) Take 200 μl of the above samples, strong positive control, critical positive control, negative control and quantitative reference standard in a 1.5ml centrifuge ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com