trail membrane-penetrating peptide-like mutant mur6, preparation method and application
A mutant and membrane-penetrating peptide technology, which is applied in the field of preparation of TRAIL membrane-penetrating peptide-like mutant MuR6, can solve the problems of low possibility of new drugs, existence of toxic and side effects, and inconspicuous advantages, so as to promote aggregation and have little impact on protein structure , enhance the effect of transduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Sequence and primer design of TRAIL penetrating peptide-like mutant
[0059] Selectively transform the 114th to 119th amino acid coding sequence of the extracellular segment of the TRAIL wild-type protein from VRERGP to RRRRRR, that is, the 114th position is mutated from valine to arginine, and the 116th position is mutated from glutamic acid to sperm Amino acid, the 118th position is mutated from glycine to arginine, the 119th position is mutated from proline to arginine, and there are 4 mutation sites, making the N-terminal of the mutant protein a coding sequence of 6 consecutive arginines , becoming a protein containing a penetrating peptide-like structure.
[0060] The cDNA encoding the mutant is shown as SEQ ID NO:1, and the amino acid of the mutant is shown as SEQ ID NO:2.
[0061] Primers were synthesized as follows:
[0062] The upstream primer MuR6-TR-NdeI is shown in SEQ ID NO: 3;
[0063] The downstream primer TR-Eco-R is shown in SEQ ID NO:4.
Embodiment 2
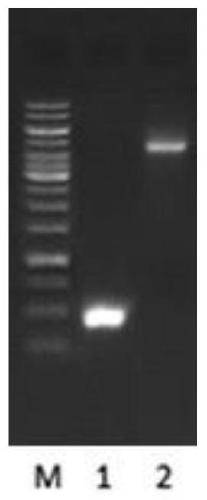
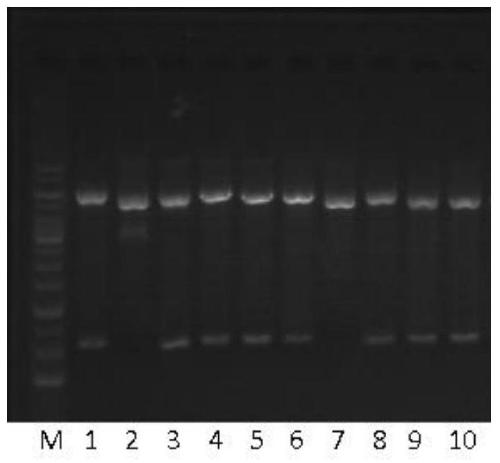
[0065] The TRAIL-MuR6 fragment was amplified by PCR and ligated with pET32a, and the single colony picking and identification of the ligated product
[0066] The TRAIL-MuR6 fragment was amplified by PCR mutation using the pMD19 / TRAIL plasmid as a template. The target fragment of TRAIL-MuR6 and the vector pET32a were digested with NdeI and EcoRI respectively. The TRAIL-MuR6 fragment was connected with the vector pET32a with the excised Trx fusion tag sequence, transformed into Top10 competent cells, single clones were picked, and identified by double digestion with XbaI and EcoRI. See Example 1 for primer design. The original sequence of the pMD19 / TRAIL plasmid was derived from NCBI Reference Sequence: NM_003810.3, and the vector pET32a was derived from Invitrogen.
[0067] Experimental procedure
[0068] 1. PCR amplification of TRAIL-MuR6 target fragment
[0069] 1. Using the pMD19 / TRAIL plasmid as a template and the MuR6-TR-NdeI / TR-Eco-R primer pair to amplify the TRAIL-Mu...
Embodiment 3
[0128] pET32a / TRAIL-MuR6 expression test
[0129] The plasmid with correct sequencing obtained in Example 2 was transformed into competent Escherichia coli BL21(DE3), and a single bacterium was picked for expression test to investigate the expression effect.
[0130] Experimental procedure
[0131] 1. Plasmid transformation and strain preservation
[0132] 1. Prepare 100ml of LB medium and sterilize at 121°C for 20min.
[0133] 2. Add 1 μl of pET32a / TRAIL-MuR6 plasmids to 100 μl of BL21(DE3) competent cells, and ice-bath for 30 minutes.
[0134] 3. Heat shock in a water bath at 42°C for 90 seconds.
[0135] 4. Incubate on ice for 3 minutes.
[0136] 5. Take 20 μl of transformed competent cells and smear them all on LB solid medium containing Amp, culture at 37°C overnight.
[0137] 6. After colonies grow on the plate the next day, pick a single bacterium on the plate and add 50ml LB (Amp + ) at 37°C overnight.
[0138] 7. Preserve 20 glycerol bacteria, the final concent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com