Sample preparation and treatment method for researching metaproteomics of soil
A technology of sample preparation and processing method, which is applied in the field of whole protein extraction and processing, can solve the problems of complex matrix, low protein content, protein extraction interference, etc., and achieve the effects of simplified operation steps, reliable and easy operation, and scientific preparation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
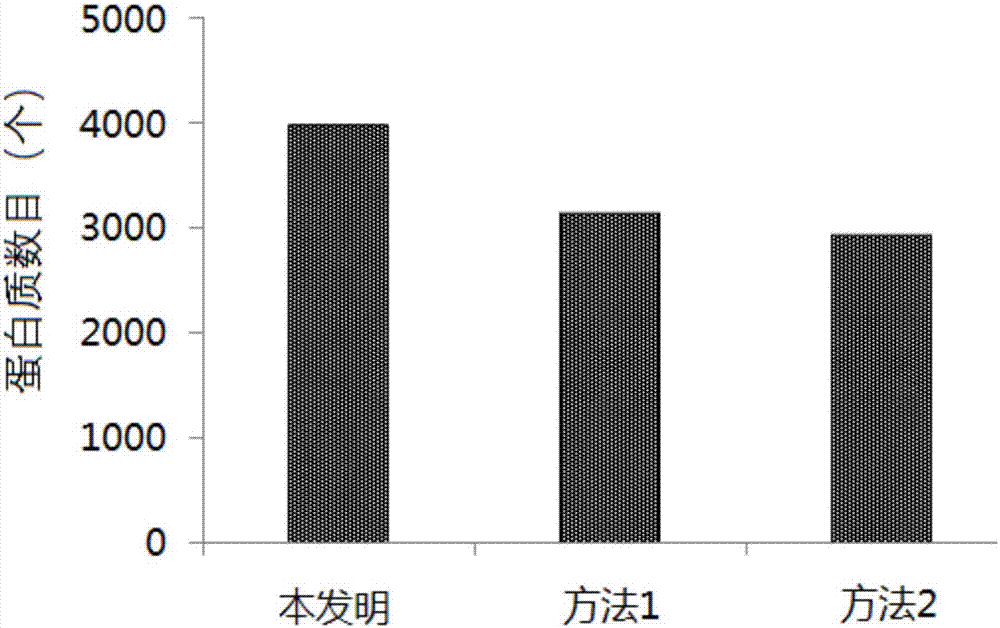
[0038] Example 1: A preparation and processing method for metaproteomics research samples in common soil samples
[0039] (1) Sample pretreatment: Put the soil sample into a freeze dryer, dry it to constant weight, and grind it in a liquid nitrogen environment. Accurately weigh 200mg of soil powder into a centrifuge tube, add 2mL of lysate, pipette with a pipette to mix well, then bathe in 95°C water for 30min, and vortex for 5min. Wherein, the formula of the lysate is: 70mM tetraethylammonium bromide (tetraethyl-ammoniumbromide, TEAB), 0.5% sodium diethyldithiocarbamate (SDC), 10mM tris (2-carboxyethyl) phosphine (Tris (2 -carboxyethyl)phosphine, TCEP), 30mM dichloroacetamide (2-chloroacetamide), 0.5% sodium dodecyl sulfate (SDS), 1M urea (Urea), the pH value was adjusted to 8.0.
[0040] (2) Vibration crushing: Add 6 steel balls with a diameter of 3mm after cleaning and baking at 400°C to the suspension obtained in step (1), vibrate and crush at low temperature, the vibrati...
Embodiment 2
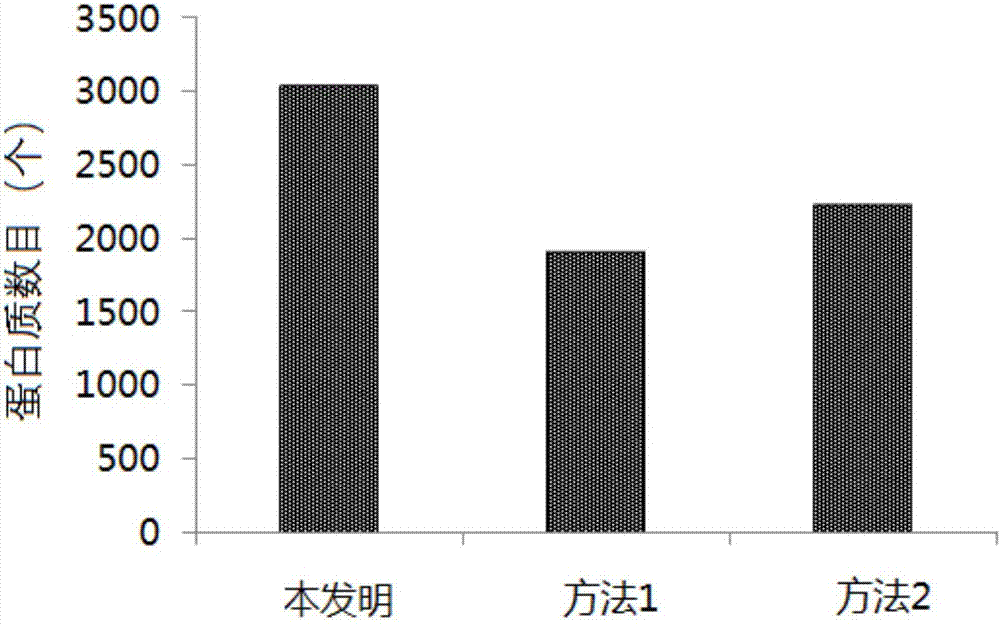
[0067] Example 2: A method for preparing and processing samples for metaproteomics research in sediment samples
[0068] (1) Sample pretreatment: Put the sediment sample (Sediment, any particle that can be moved by fluid flow, and eventually become a layer of solid particles under water or other liquids) into a freeze dryer and dry to constant weight , ground under liquid nitrogen environment. Accurately weigh 300mg of soil powder into a centrifuge tube, add 3mL of lysate, pipette with a pipette to mix well, then bathe in 90°C water for 1 hour, and vortex for 5min. The formula of the lysate is: 70mM tetraethylammonium bromide (tetraethyl-ammoniumbromide, TEAB), 0.5% sodium diethyldithiocarbamate (SDC), 10mM tris (2-carboxyethyl) phosphine (Tris (2-carboxyethyl) )phosphine, TCEP), 30mM dichloroacetamide (2-chloroacetamide), 0.5% sodium dodecyl sulfate (SDS), 1M urea (Urea), the pH value was adjusted to 8.2.
[0069](2) Vibration crushing: Add 6 steel balls with a diameter of ...
Embodiment 3
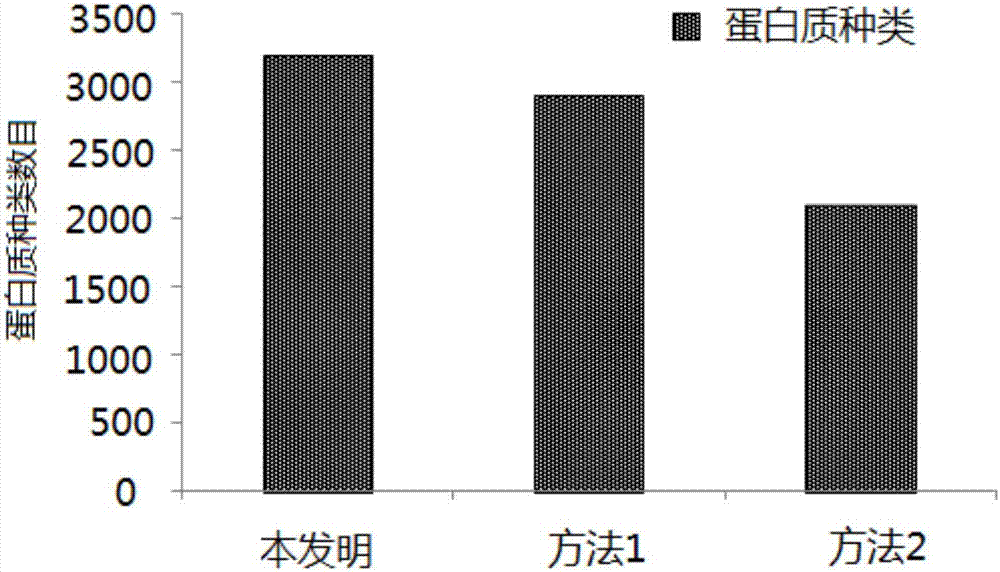
[0096] Example 3: A method for preparing and processing samples for metaproteomics research in sludge samples
[0097] (1) Sample pretreatment: Put the sludge sample into a freeze dryer, dry it to constant weight, and grind it in a liquid nitrogen environment. Accurately weigh 400 mg of sludge powder into a centrifuge tube, add 4 mL of lysate, and pipette to mix well, then bathe in 80°C water for 2 hours, and vortex for 5 minutes. Wherein, the formula of the lysate is: 70mM tetraethylammonium bromide (tetraethyl-ammoniumbromide, TEAB), 0.5% sodium diethyldithiocarbamate (SDC), 10mM tris (2-carboxyethyl) phosphine (Tris (2 -carboxyethyl)phosphine, TCEP), 30mM dichloroacetamide (2-chloroacetamide), 0.5% sodium dodecyl sulfate (SDS), 1M urea (Urea), the pH value was adjusted to 8.0.
[0098] (2) Oscillation crushing: Add 8 steel balls with a diameter of 3mm after cleaning and baking at 400°C to the suspension obtained in step (1), and oscillate and crush at low temperature, the ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com