Cleavage efficiency enhanced substrate mutant of HRV (Human Rhinovirus) 3C protease and application thereof
A protease substrate, variant technology, applied in the field of bioengineering, can solve the problems of difficult screening, large workload, low efficiency, etc., and achieve the effect of efficient cutting advantages
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
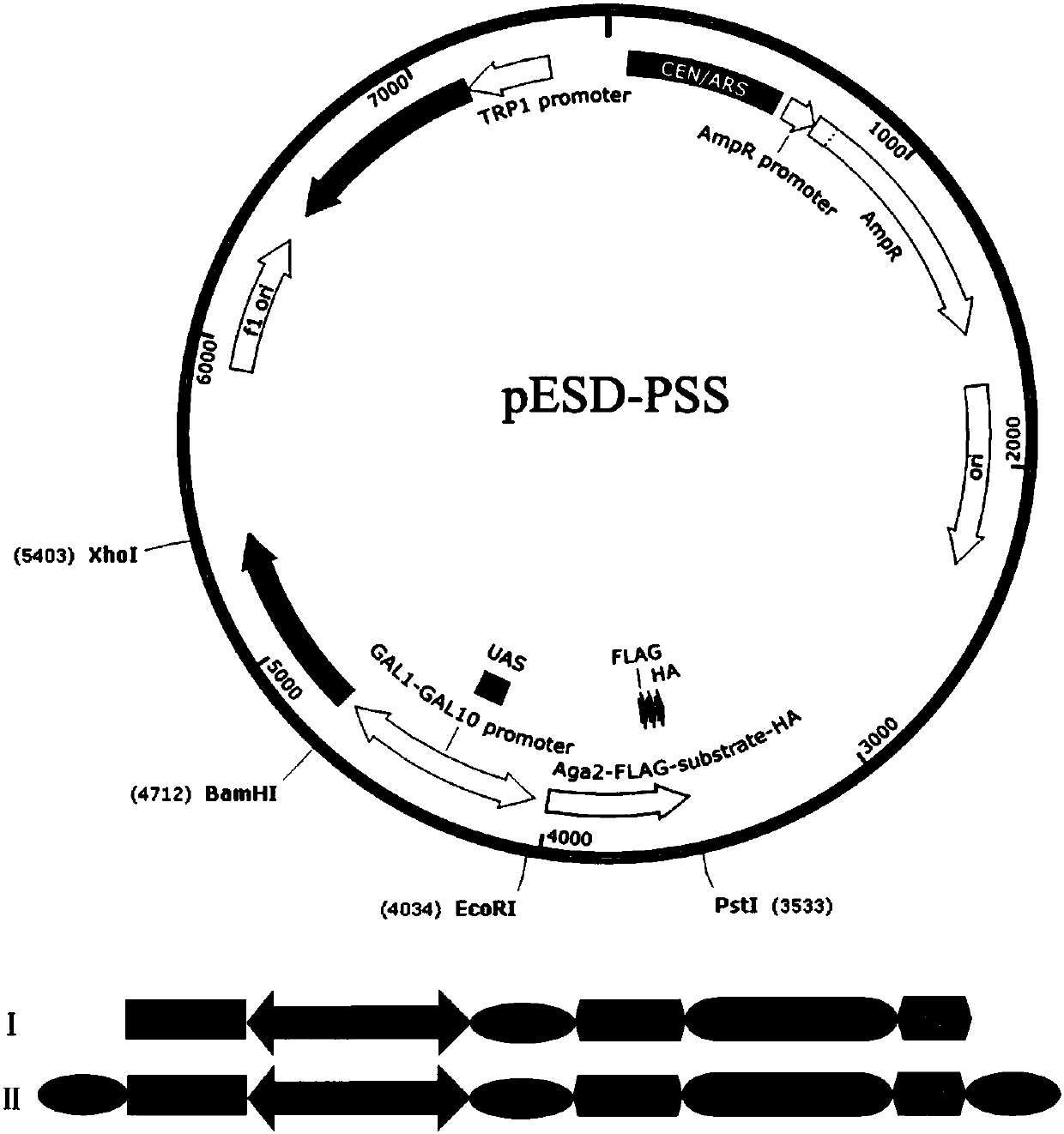
[0022] Example 1 Construction of HRV 3C protease and its substrate mutants inducible expression plasmids in Saccharomyces cerevisiae
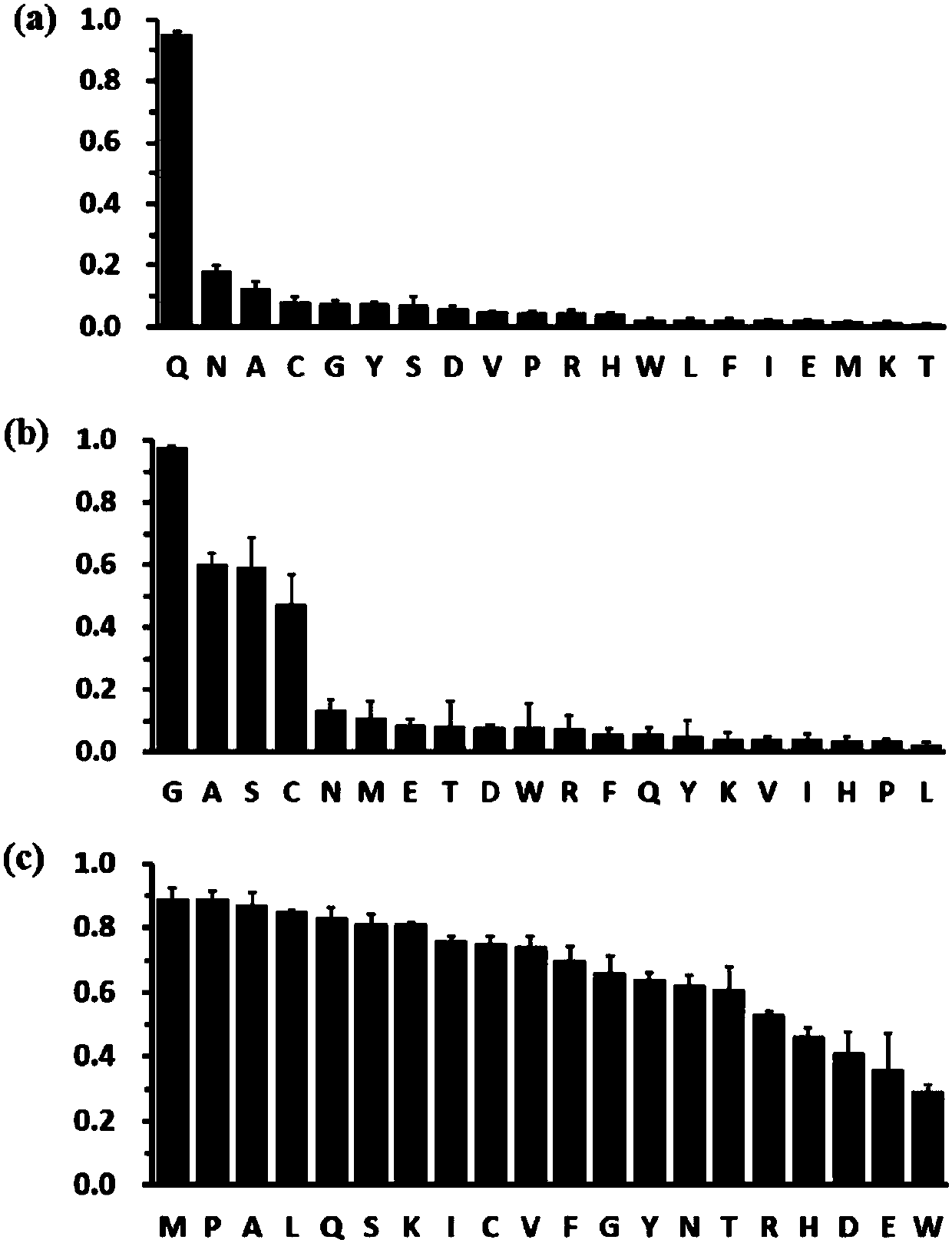
[0023] (1) The substrate mutant gene fragment was amplified by three rounds of polymerase chain reaction (PCR). Among them, site-directed mutagenesis was carried out in the first round of PCR amplification to obtain the mutant substrate genes of HRV 3C protease substrate cleavage sites P1, P1', P2'; the second round of PCR amplification was used to determine whether the gene fragment had a retained signal peptide gene ; In the third round of PCR amplification, the restriction endonuclease cuts the gene sequence to obtain the complete gene. Among them, there are 20 kinds of amino acid saturation mutants with stranded signal peptide genes at the P1 site, and 3 kinds of mutant genes with amino acids Q, N, and F without the stranded signal peptide genes; There are 20 kinds of amino acid saturation mutants with stranded signal peptide genes, and 3 ...
Embodiment 2
[0031] Example 2 Induced expression of plasmids containing HRV 3C protease and substrate mutants thereof in Saccharomyces cerevisiae
[0032] Transform the final vector into Saccharomyces cerevisiae EBY100, and the reagents used for transformation are: EBY100 competent cells, 20 μL; recombinant plasmid, 1.5 μL; Single strand carrier DNA, 25 μL; 1M LiAC, 36 μL; PEG4000 (50% W / V), 240 μL After mixing the system, put it in a water bath at 30°C for 30 minutes, transfer it to 42°C for heat shock for 20 minutes, collect the bacteria by centrifugation, add 1mLYPD liquid culture and culture on a shaker at 30°C for 1.5hr, wash the cells once with ultrapure water, and then spread them on YNB- CAA-Glucose solid medium, cultivated in a 30°C incubator for 2-3 days. The composition of YNB-CAA-Glucose solid medium is 20g / L glucose, 6.7g / L YNB, 5.4g / L Na 2 HPO 4 ,8.6g / L NaH 2 PO 4 ·H 2 O and 5g / L casamino acids, pH7.4, 15g / L agar.
[0033] Inoculate a single colony in the liquid medium ...
Embodiment 3
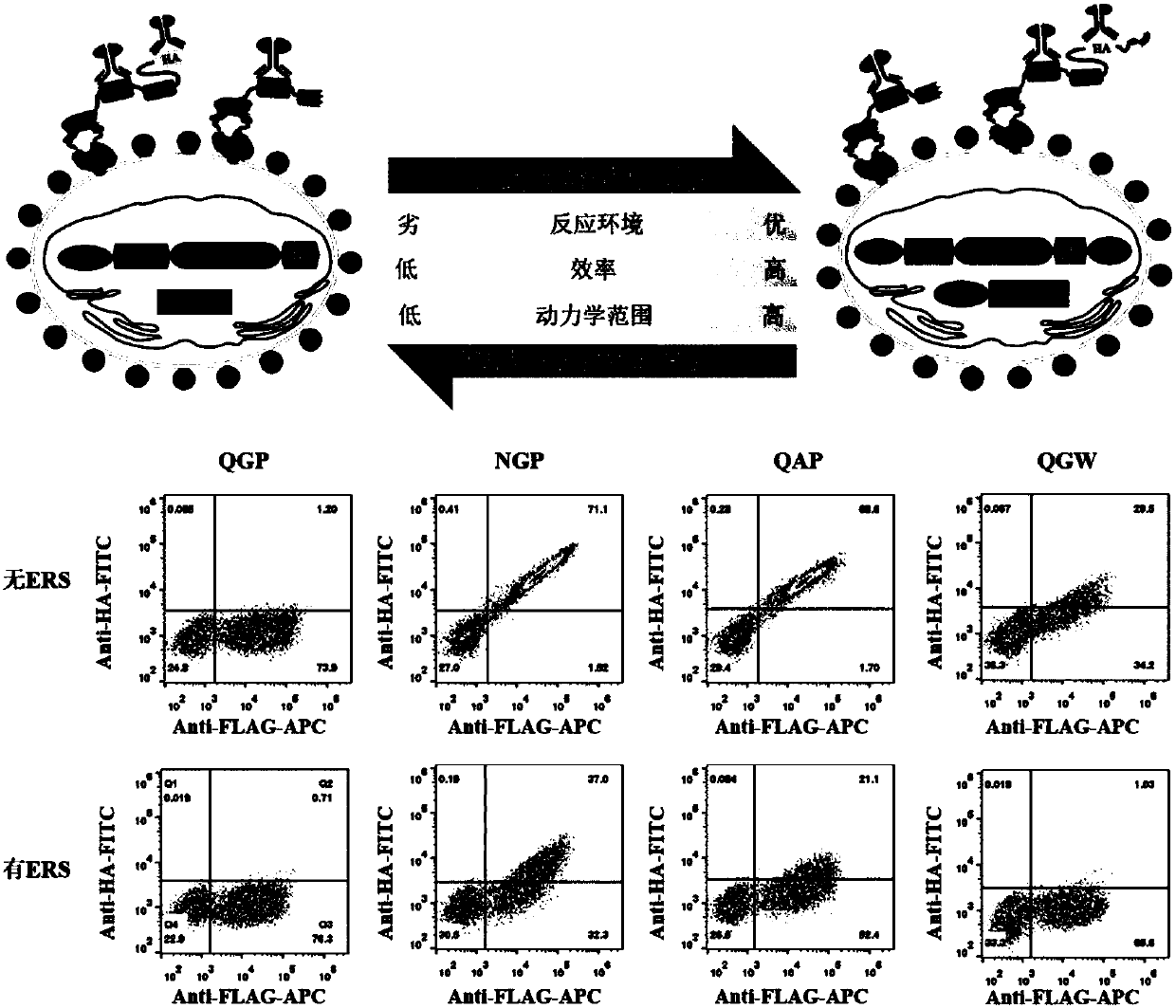
[0034] Example 3 Application of flow cytometry to detect the reaction results of HRV 3C protease and its substrate mutants
[0035] take 10 6 Add 200 μL of Buffer A to mix well, then centrifuge to pellet the cells, 3000 rpm, 4°C, 1 min. Then wash the cells with Buffer B solution, remove the supernatant, then add 20 μL Buffer B solution, 0.15 μL FITC, 0.15 μLAPC to each sample, label in the dark at 4°C for 15 minutes, and transfer to room temperature for 30 minutes in the dark. Centrifuge to remove the supernatant, then wash once with 150μL Buffer B, continue to wash once with 1x PBS, and resuspend with 300μL. The resuspended cells were analyzed by CytoFLEX flow cytometer, and the fluorescent signal channels detected were FITC (525 / 40nm) and APC (660 / 20nm). The cleavage efficiency of HRV 3C protease to its substrate mutants was obtained from the analysis of the results given in the flow cytometer (see image 3 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com