Amplification primer for detecting PAH (phenylalanine hydroxylase) gene mutation, kit and detection method thereof
A detection method and amplification primer technology, which can be used in biochemical equipment and methods, recombinant DNA technology, and microbial determination/inspection, etc. The detection rate and the effect of simplifying the experimental operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Genomic DNA extraction: collect 2ml of peripheral blood from the subject, put it in an EDTA anticoagulant tube, take 0.2ml of EDTA anticoagulant peripheral blood sample, extract DNA according to the instructions of the whole blood DNA extraction kit, and analyze the DNA concentration with Qubit2.0. The extracted genomic DNA is ready to be used as a PCR template for the next step. In this study, a total of 5 genetic DNA samples were tested, of which 1 genetic DNA sample had known pathogenic mutation sites, and 4 were normal human genomic DNA.
Embodiment 2
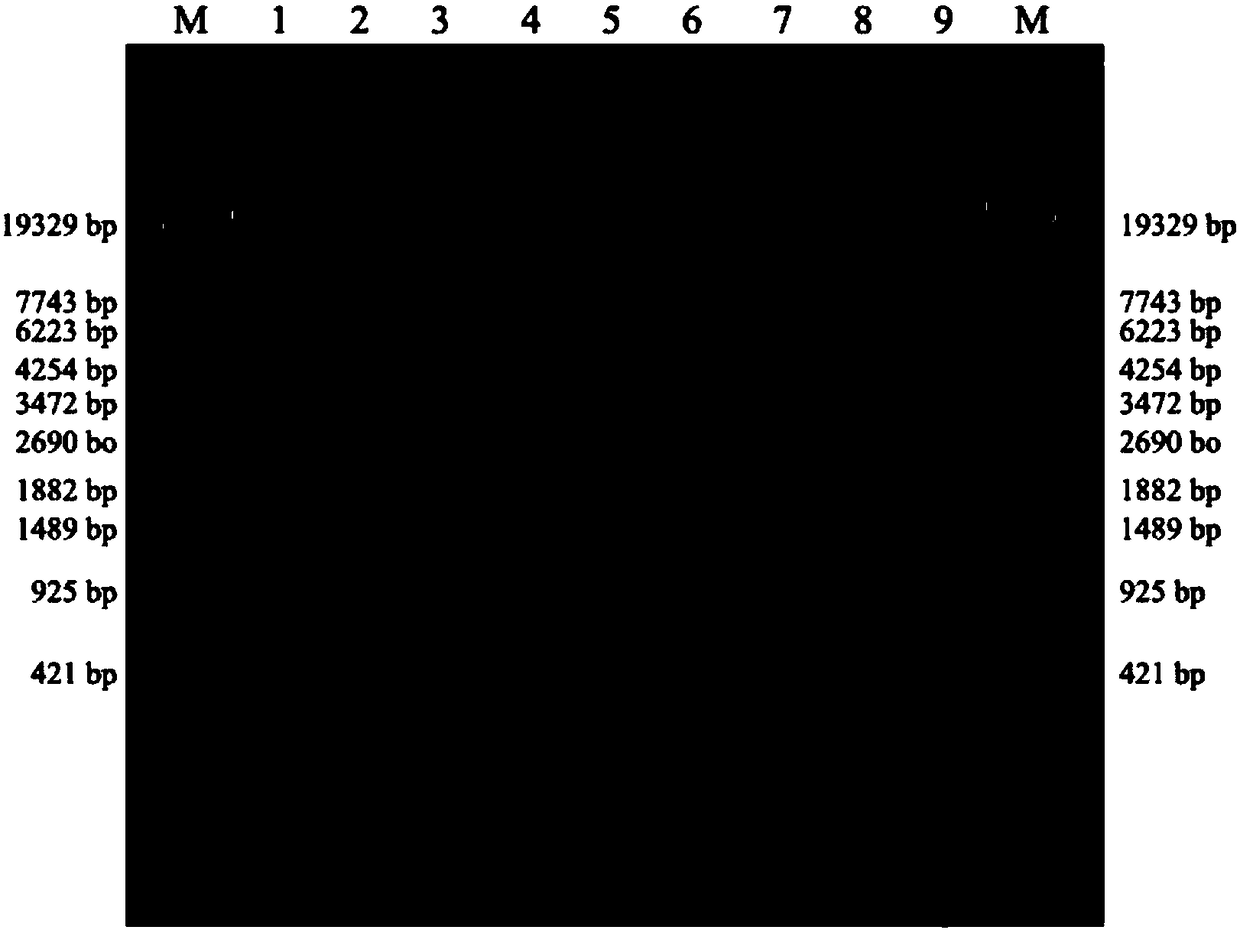
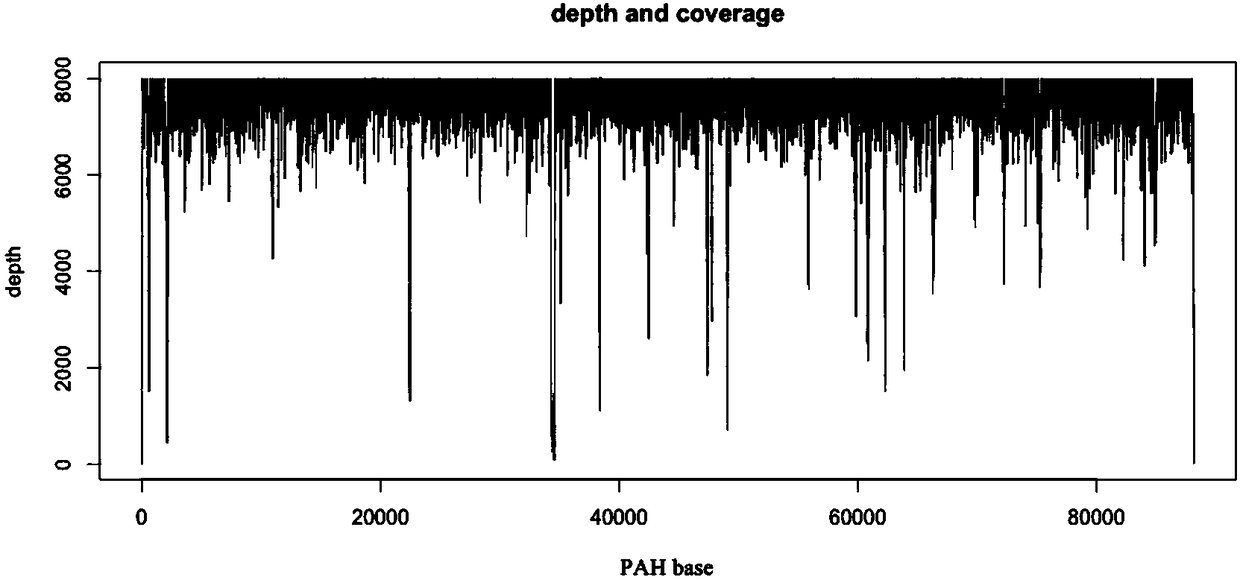
[0049] LR-PCR amplification: According to the PAH gene sequence, 9 pairs of PAH gene-specific primers were designed and synthesized, and GeneAmp HighFidelity PCR System was used for LR-PCR to amplify 9 large fragment sequences, respectively 1, 2, 3, 4, 5, 6, 7, 8 and 9, the primer sequences and amplified product sizes are shown in Table 1, using the GeneAmp High Fidelity PCR System, setting the reaction conditions and performing LR-PCR, PCR on a Veriti 96-well Thermal Cycler PCR instrument The total reaction volume is 50 µl, including 5 µl of 10× GeneAmp High Fidelity PCR buffer, 2 µl of 10 mmol / L dNTP, 1 µl of 10 µmol / L forward and reverse primers, 5 µl of 5 mol / L betaine, 2.5 µl of DMSO, 5 U / µL for polymerization Enzyme mixture 1μl, 50ng / μl genomic DNA 2μl, water 30.5μl, PCR cycle parameters: denaturation at 98°C for 1min; 98°C for 10sec, 68°C for 13min, a total of 30 cycles, and finally 72°C for 15min, PCR reaction in Veriti 96-well Thermal Completed on a Cycler PCR instrum...
Embodiment 3
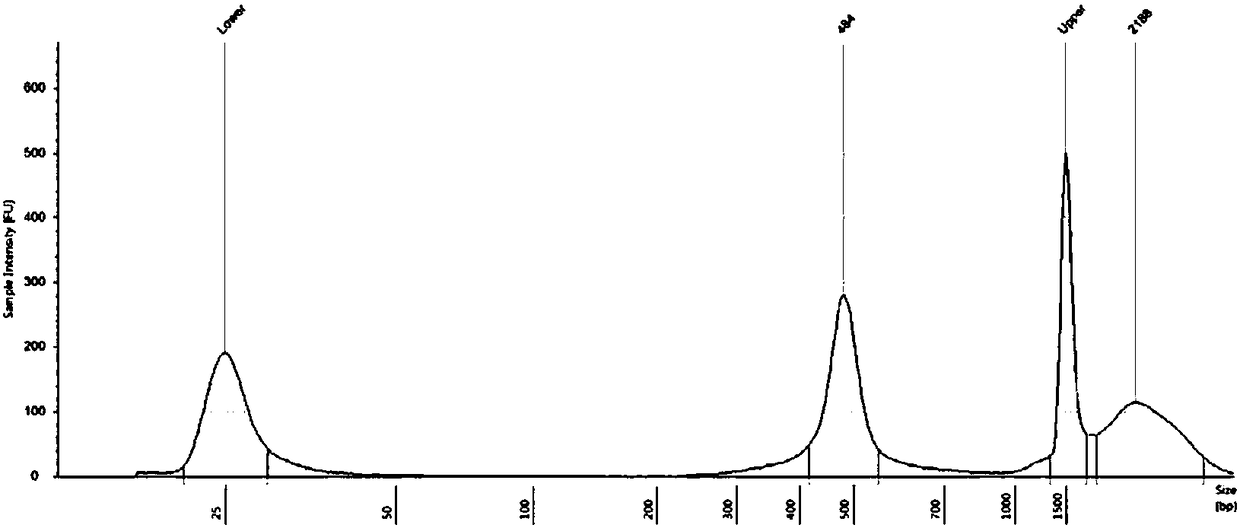
[0055] Purification and quantification of LR-PCR products: LR-PCR products were purified by the magnetic bead method, and the DNA concentration of No. 1 to No. 9 purified products of each genomic DNA sample was quantified using a Qubit 2.0 fluorescence photometer. The concentration detection results of 45 LR-PCR amplification products are as follows:
[0056]DNA concentration detection (Qubit 2.0)
[0057]
[0058] Since the lengths of the nine amplified fragments are basically the same, the nine PCR products corresponding to the same genomic DNA sample were mixed according to equal mass, and concentrated and enriched with Agencourt AMPure XP magnetic beads (Beckman Coulter, USA). 2.0 fluorometer for quantification.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com