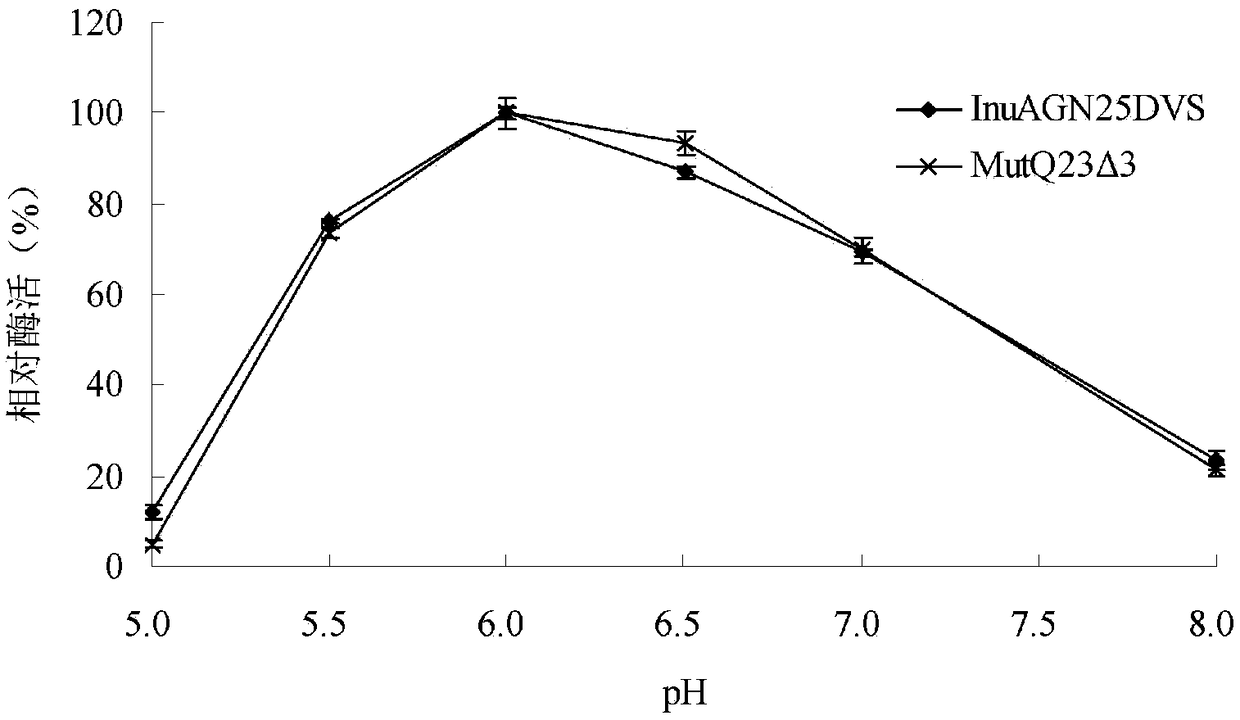
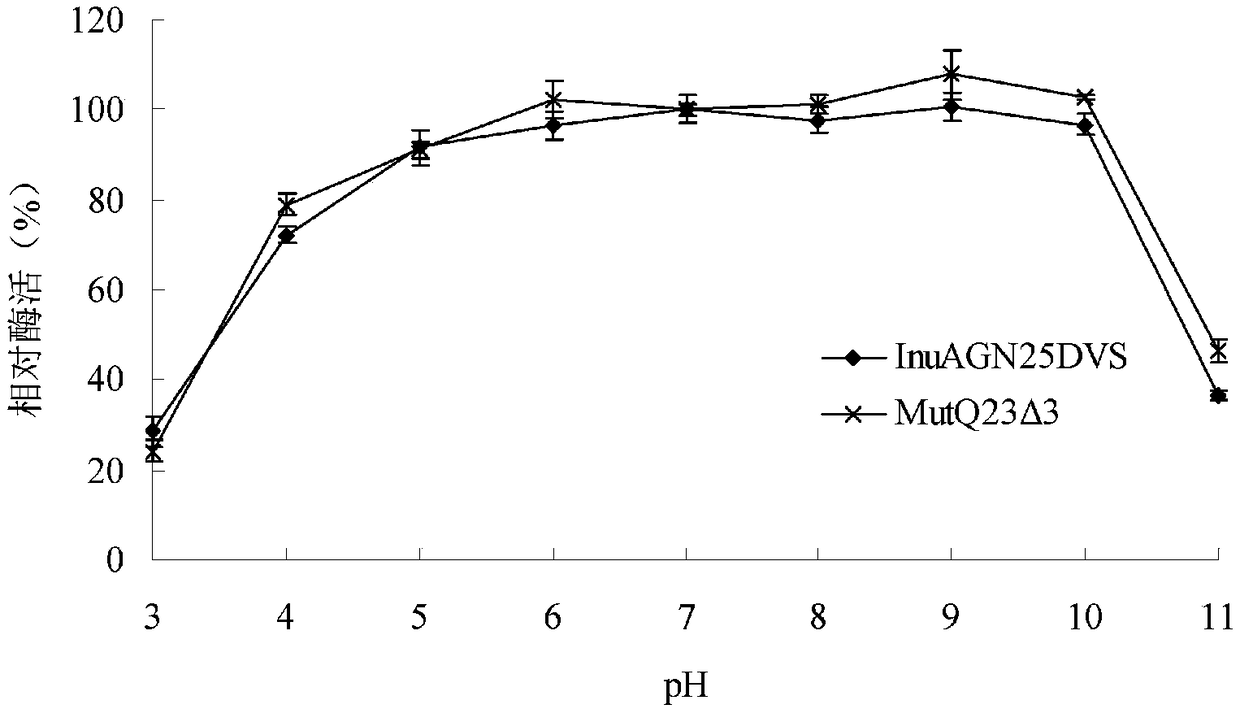
Heat-resistant low temperature exo-inulinase mutant MutQ23 delta 3
An exo-inulinase and mutant technology, which is applied in the field of genetic engineering, can solve the problems of poor heat resistance of low-temperature enzymes, and achieve the effect of improving thermal stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Construction and Transformation of Embodiment 1 Wild Enzyme InuAGN25DVS Expression Vector
[0036] 1) According to the exo-inulinase nucleotide sequence JQ863108 (SEQ ID No.4) recorded in GenBank, design primers 5'TGGAAGTTCTGTTCCAGGGGCCCCAGACGGGACAGCATAAACAAG 3' and 5'GGTGGTGGTGTTATCTCTTAAATGCAGAAATACCGAT 3', and use the plasmid pEASY-E1-Z2-5 as a template PCR amplification, the exo-inulinase mature peptide coding sequence Z2-5 was obtained, and the HRV 3C protease cleavage site coding sequence was added at the 5' end of Z2-5, and at the 5' end of Z2-5 and the 3 ' end also formed a recombination region that matched the recombination region at both ends of the linearized vector obtained in (2). Z2-5 can also be obtained by gene synthesis.
[0037]2) Also design primers 5'AGAGATAACACCACCACCACCACCACTG 3' and 5'CCTGGAACAGAACTTCCAGGAATTCGGATCCGCGACC 3', and use the pET-28a(+) plasmid as a template for PCR amplification to obtain a linearized pET-28a(+) vector. The 5' and 3...
Embodiment 2
[0042] Example 2 Construction and transformation of mutant enzyme MutQ23Δ3 expression vector
[0043] 1) Synthesize the MutQ23Δ3 coding gene sequence, add HRV 3C protease restriction site coding sequence and EcoRI restriction enzyme cutting site sequence (5'GAATTCCTGGAAGTTCTGTTCCAG 3') at the 5' end of the coding gene during synthesis, and add The XhoI restriction site sequence (5'CTCGAG 3') was added to the 3' end.
[0044] 2) The sequence synthesized in (1) is subjected to EcoRI and XhoI double digestion; at the same time, the expression vector pET-28a(+) is subjected to EcoRI and XhoI double digestion.
[0045] 3) Ligate the digested product in (2) with DNA ligase to obtain the expression vector of the mutant enzyme MutQ23Δ3.
[0046] 4) The ligation product was transformed into Escherichia coli BL21(DE3) to obtain a recombinant strain containing the gene encoding MutQ23Δ3.
Embodiment 3
[0047] Example 3 Preparation of Wild Enzyme InuAGN25DVS and Mutant Enzyme MutQ23Δ3
[0048] Recombinant strains containing Z2-5 and MutQ23Δ3 coding genes were inoculated in LB (containing 50 μg mL -1 Kanamycin) medium, shake rapidly at 37°C for 16h.
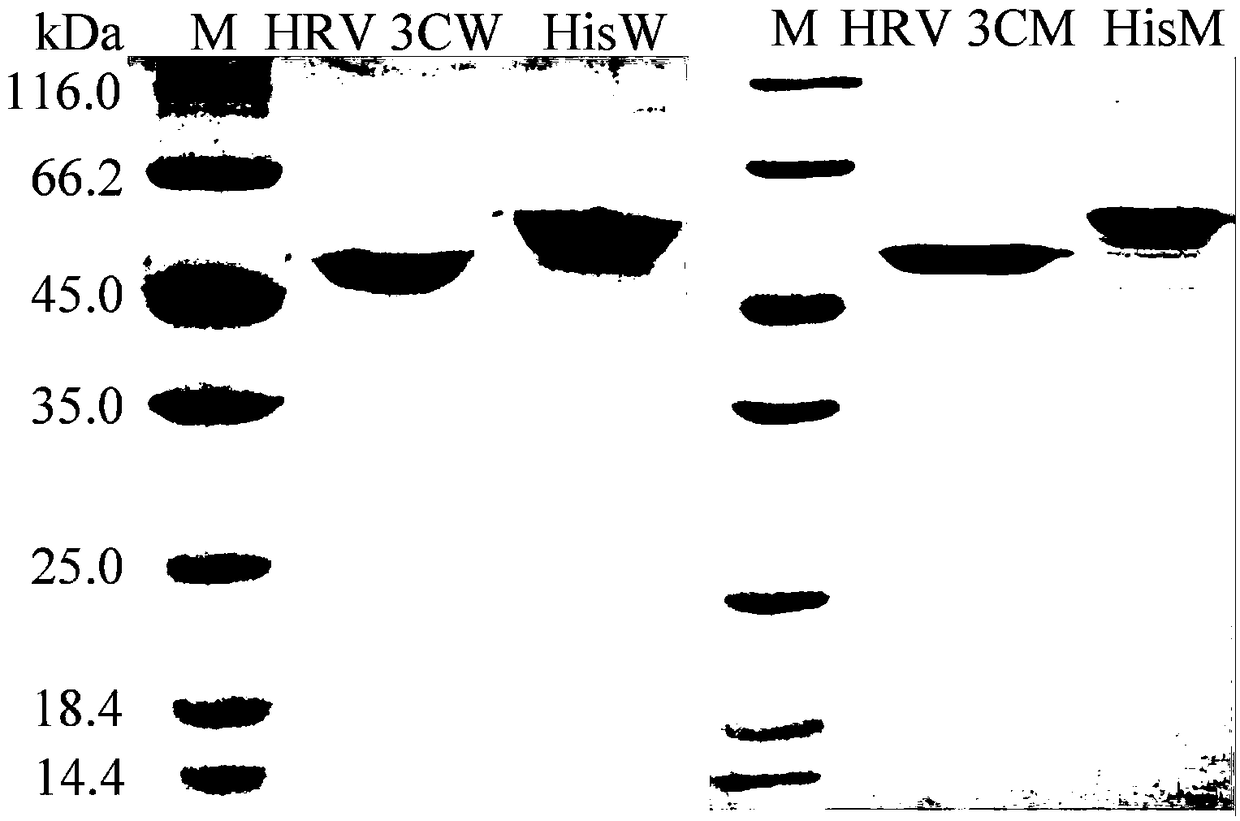
[0049] Then inoculate the activated bacterial solution into fresh LB (containing 50 μg mL -1 Kanamycin) medium, rapid shaking culture for about 2–3h (OD 600 After reaching 0.6-1.0), add IPTG with a final concentration of 0.25mM for induction, and continue shaking culture at 20°C for about 20h. Centrifuge at 12000rpm for 5min to collect the bacteria. After suspending the cells with an appropriate amount of McIlvaine buffer with pH=7.0, the cells were ultrasonically disrupted in a low-temperature water bath. The crude enzyme solution concentrated in the cells above was centrifuged at 13,000rpm for 10min, the supernatant was aspirated and the target protein was affinity and purified with Nickel-NTA Agarose and 0-500mM imidazole ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com