Fidaxomicin genetically engineered bacterium, construction method and application
A technology of genetically engineered bacteria and fidaxomicin, which is applied in the field of high-yielding fidaxomycin genetically engineered bacteria Actinomycetes Deccan plateau YP-2 and its construction, and achieves high efficiency, reduced production cost, and quick and convenient operation. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Construction of a genetically engineered bacterium that produces high-yield fidaxomicin Actinoplanes deccanensis YP-2 (Actinoplanes deccanensis YP-2)
[0037] 1. Construction of pathway-specific gene expression vector pIJ8630-ermE*-fadR1
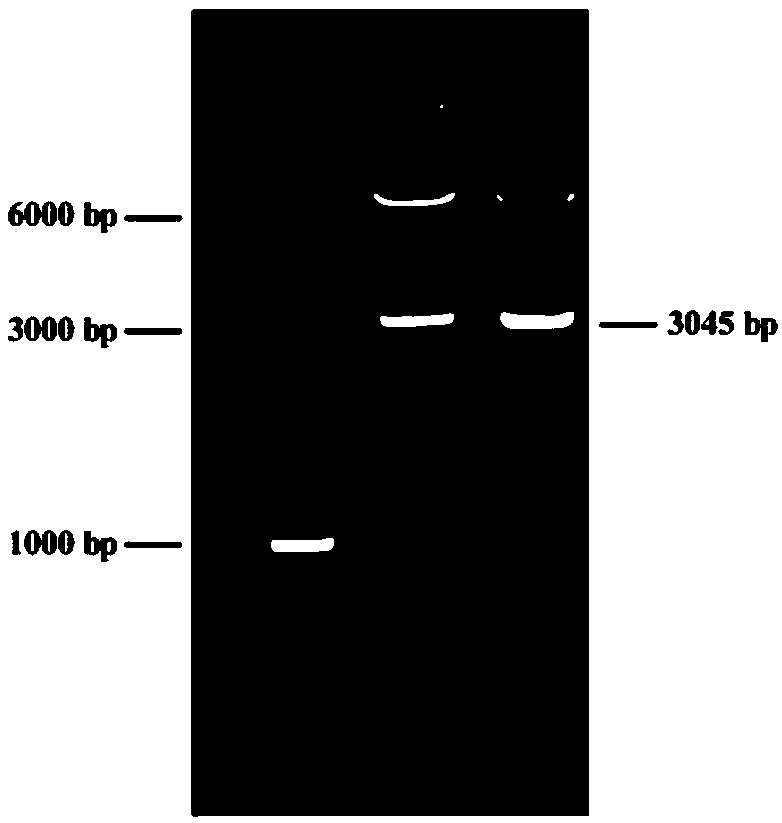
[0038] The name of the expression vector of the positive regulatory gene of fidaxomicin biosynthesis constructed in this example is pIJ8630-ermE*-fadR1, which contains the positive regulatory gene fadR1 of fidaxomicin biosynthesis. The positive regulatory gene of fidaxomicin biosynthesis The sequence of fadR1 is shown in SEQ ID NO.1. SEQ ID NO.1 consists of 3063 nucleotides, the 1st-19th is the NdeI recognition site and the protective base, the 20th-297th is the erythromycin resistance gene promoter, and the 297th-3042 is Feida The coding sequence of the positive regulation gene of biosynthesis of Fidaxomycin, the 3043-3063 position is the NdeI recognition site and the protection base, and SEQ ID NO.2 is the amino acid sequ...
Embodiment 2
[0048] Example 2 Fermentation verification of Fidaxomycin synthesized by starting bacteria and genetically engineered strains.
[0049] (1) Cultivate the starting bacteria Actinoplanes deccanensis YP-1 and the genetically engineered strain Actinoplanes deccanensis YP-2 (Actinoplanes deccanensis YP-2) on ISP4 solid medium 10 days.
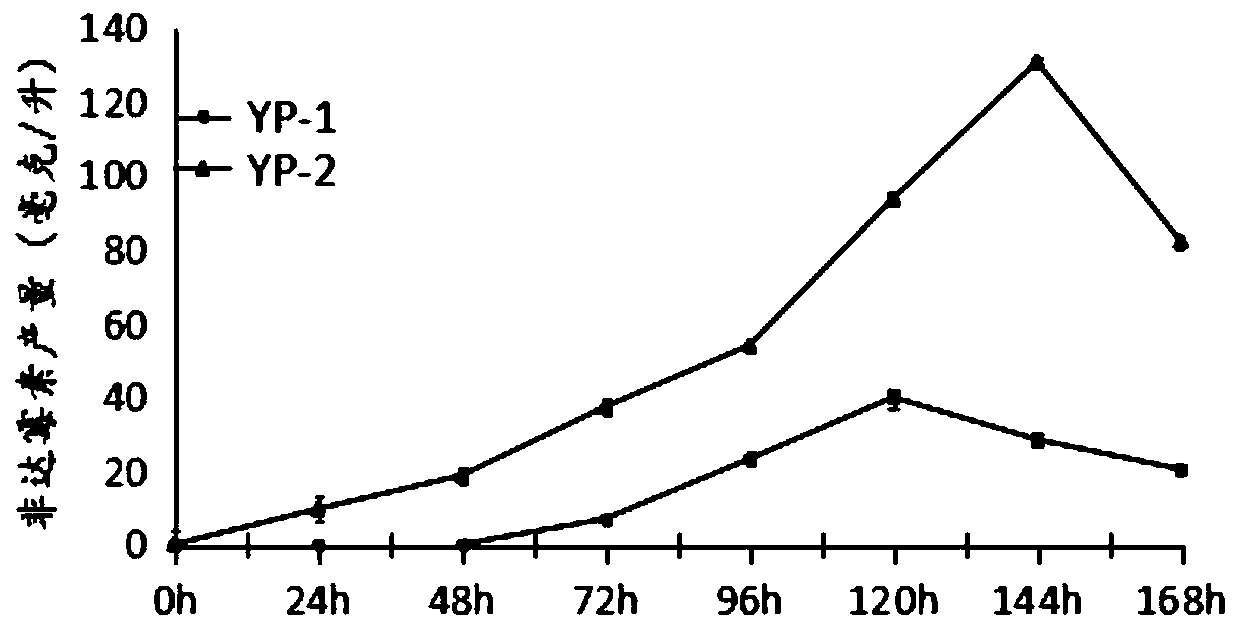
[0050] (2) On the ISP4 solid medium cultured by Actinomycetes YP-1 and Actinomyces YP-2, take 1cm×1cm pieces of bacteria and inoculate them into the seed medium medium, cultivated at 30°C for 16 hours, with a rotation speed of 200rpm; inoculated the mycelium in the seed medium into the fermentation medium B to OD 600 0.15, cultured at 30°C for 168 hours, after 12, 24, 48, 72, 96, and 120 hours of culture, samples were taken to measure the biomass of bacteria and the yield of fidaxomicin. Experiments were repeated three times.
Embodiment 3
[0051] Example 3 Genetic engineering strains and starting bacterium biomass, fidaxomicin output comparative verification
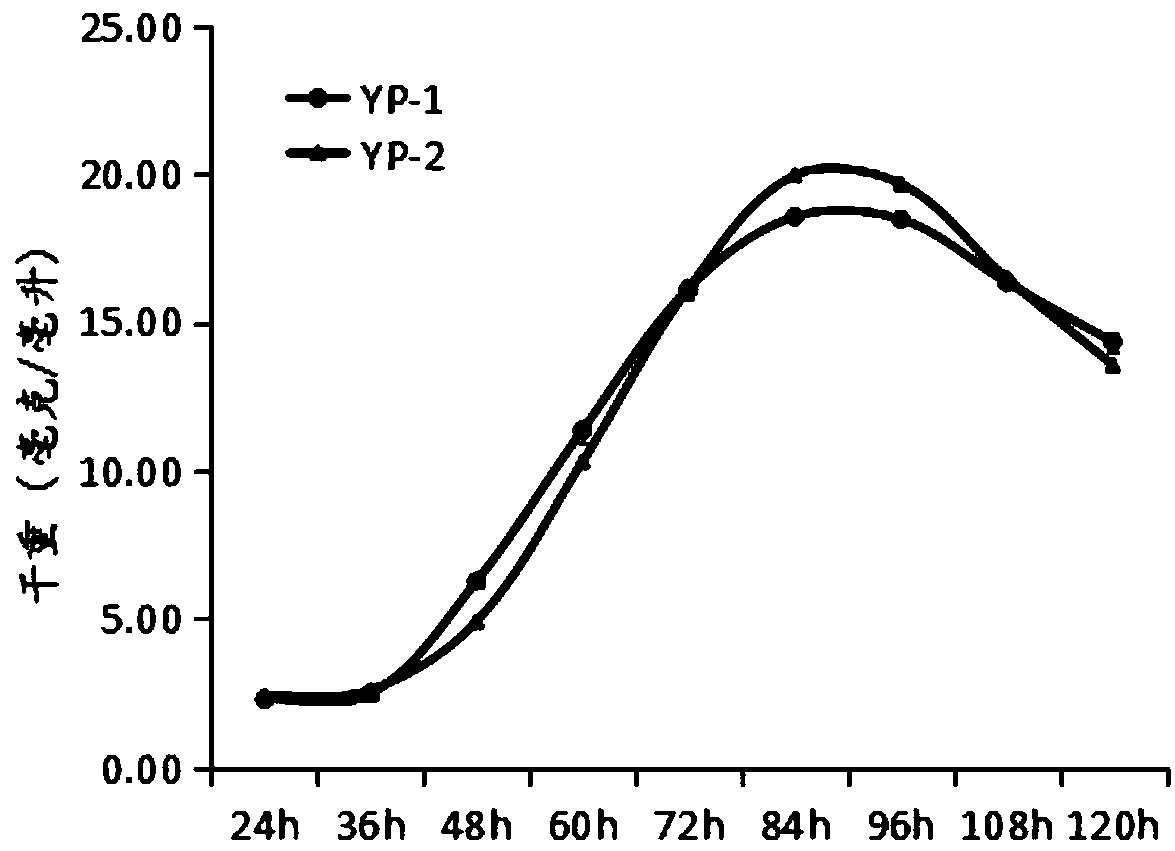
[0052] (1) Bacterial biomass: Take 1ml of the bacterial liquid fermented by fermentation medium B, collect the bacterial cells after centrifugation, wash with 1ml of sterile water and centrifuge again to collect the bacterial cells, dry at 50°C for 3 days, and then weigh them. figure 2 It is the biomass curve of Actinomycetes YP-1 and Actinomycetes YP-2.
[0053] (2) Fidaxomycin standard curve: the Fidaxomicin standard substance was prepared into a solution with a certain concentration gradient with methanol, and determined by HPLC. HPLC conditions: Chromatographic column: C18 column (Aglient, Eclipse Plus XDB, 5um, 4.6mm*250mm); detection wavelength: 254nm; flow rate: 1.00mL / min; injection volume: 10ul; experimental mobile phase: mobile phase A phase is 10% acetonitrile, containing 0.08% trifluoroacetic acid (TFA), mobile phase B phase is 90% acetonitri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com