Desaturase from arbuscular mycorrhizal fungi Delta 17 and application thereof
A saturase and unsaturated fatty acid technology, which is applied in applications, fungi, enzymes, etc., can solve the problems of limiting comprehensive catalytic activity and weak conversion rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Example 1: Determination of Δ17Des multiple sequence alignment template
[0026] Through literature research and sequence comparison of NCBI library, Δ17Des genes that currently prefer 20Cω-6LC-PUFAs were screened, and 6 genes were found to be reported (Saprolegniasis, Phytophthora infestans, Pythium melon, Sudden death of oak Phytophthora sojae, Phytophthora sojae, Phytophthora parasitica), and according to their catalytic efficiency to various substrates (see Table 1), Δ17Des (oPaFADS17) with the highest AA conversion rate was selected as the template.
[0027] Table 1 The reported catalytic efficiency of 20C-preferring Δ17Des on LA / GLA / DGLA and AA substrates
[0028]
Embodiment 2
[0029] Example 2: Δ17Des Multiple Sequence Alignment Determination of Candidate Genes
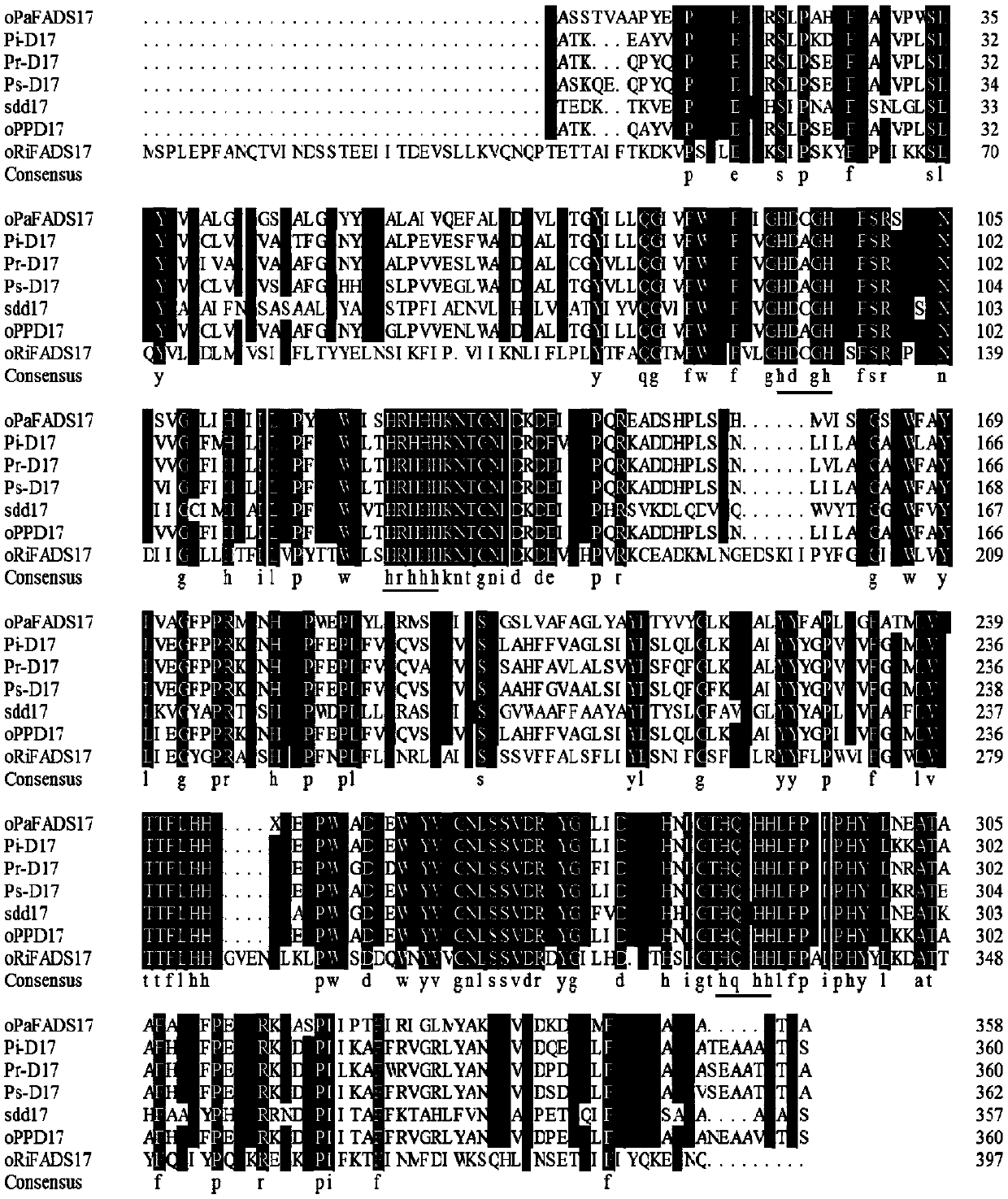
[0030] The gene sequence of Δ17 desaturase oPaFADS17, which prefers 20Cω-6LC-PUFAs, was used as a template for comparison in the NCBI library. According to the similarity and homology of the sequences, four gene sequences with high similarity and close species kinship were screened out. Combining with the host lipid content and fatty acid species of the research gene in the literature, the gene oRiFADS17 from a new species of arbuscular mycorrhizal fungi was finally determined as a candidate gene (the gene number is GBC46259.1, as shown in SEQ ID NO.2), NCBI The definition of this section of gene is acyl ω-3 fatty acid desaturase, fatty acid domain-containing protein, and endoplasmic reticulum enzyme, which are all characteristic of desaturase, but their specific functions are not described.
[0031] Using Clustal Omega software to compare the oRiFADS17 candidate gene with six 20C-preferrin...
Embodiment 3
[0034] Example 3: Construction and verification of recombinant expression vectors
[0035] 1. Codon optimization and synthesis of oRiFADS17 candidate genes
[0036] Using the sequence of oPaFADS17 as a template, the oRiFADS17 gene was determined by BLAST alignment. Because the codon preference of the gene sequence of arbuscular mycorrhizal fungi is different from that of the host Saccharomyces cerevisiae, in order to increase the expression of the target protein, we optimized the coding gene according to the codon usage of the natural sequence of the target strain, and synthesized oRiFADS17 Coding region, optimized gene sequence such as SEQ ID NO.3 (Nanjing GenScript Company, China). Their codon adaptation indices (CAI) were all raised to above 0.9 (CAI>0.8 is preferred). The synthetic sequence was ligated with the vector PUC57-simple through EcoRI and XhoI to obtain the subcloning vector PUC57-oRiFADS17, which was stored in E. coli Top 10.
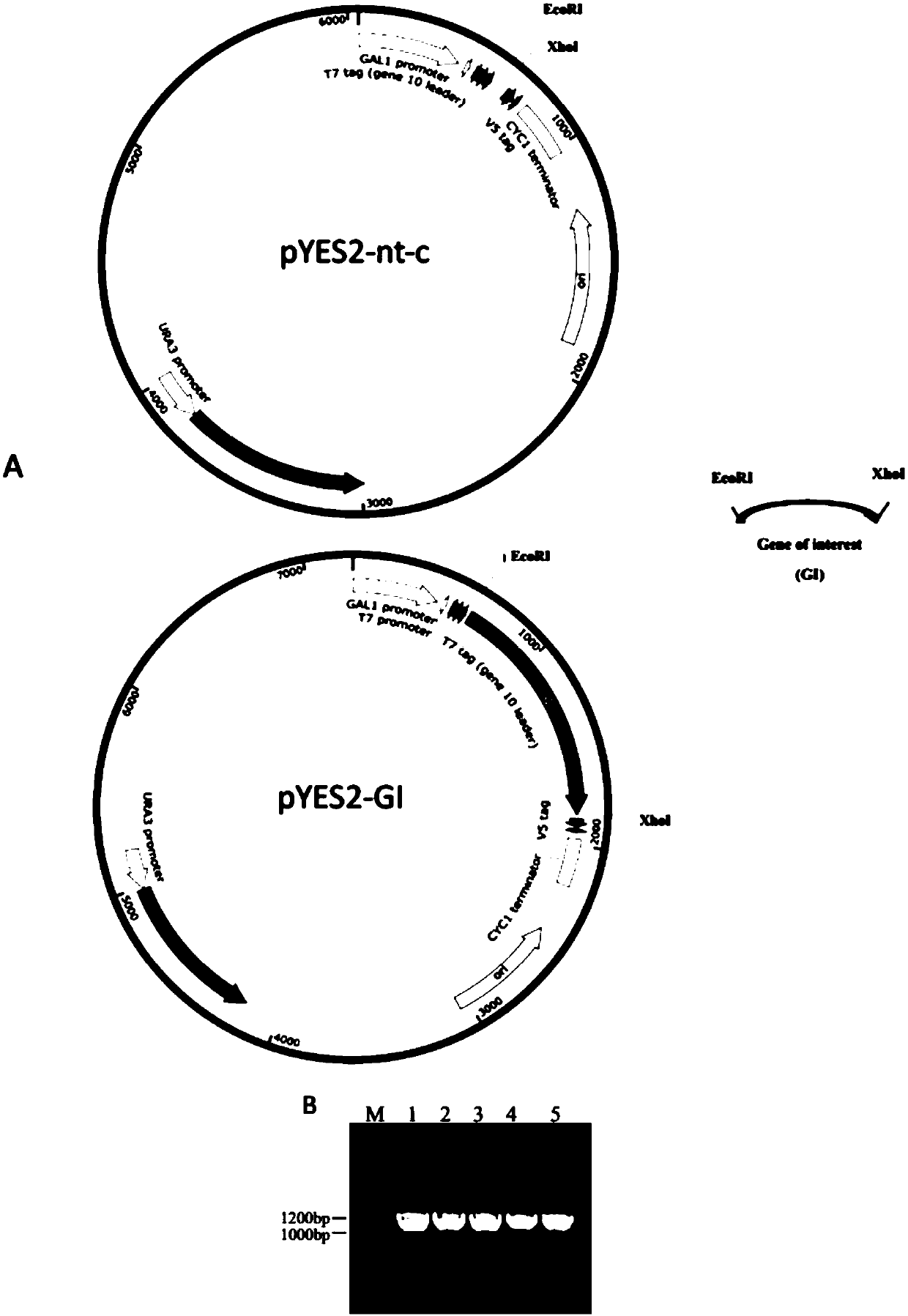
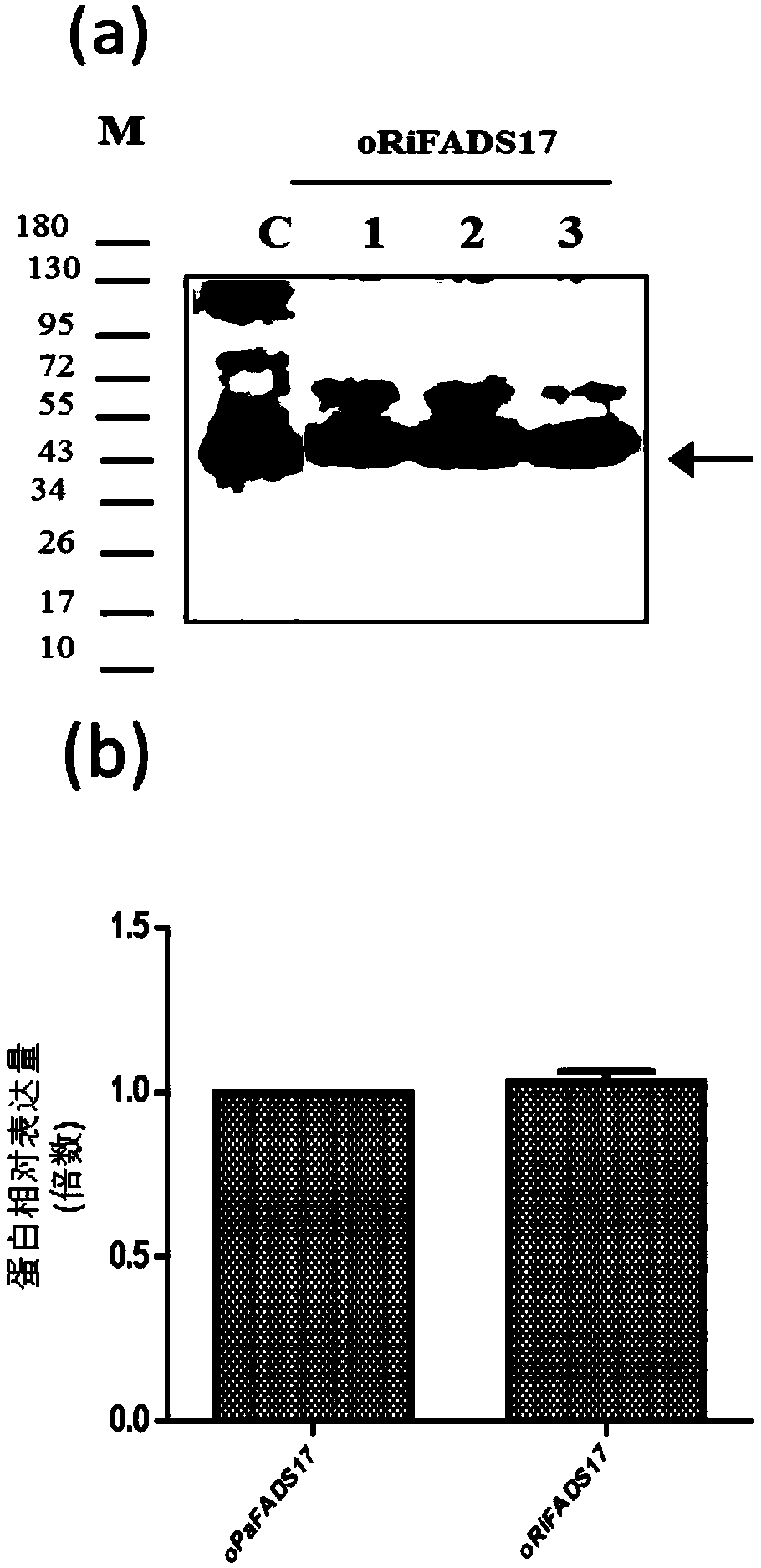
[0037] 2. Construction of recom...
PUM
| Property | Measurement | Unit |
|---|---|---|
| conversion efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com