Genetically engineered bacterium for efficiently immobilizing enterococcus faecium glutamate decarboxylase and immobilization method
A technology of genetically engineered bacteria and Enterococcus faecium, applied in the direction of microorganism-based methods, chemical instruments and methods, biochemical equipment and methods, etc., can solve the problem of low GAD expression, inability to reuse and continuous production, free enzymes Difficult to recycle and other issues, to achieve the effect of cost reduction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
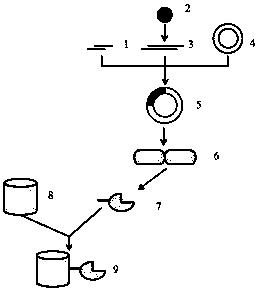
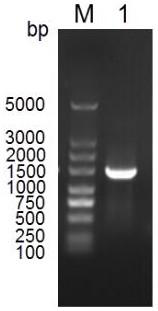
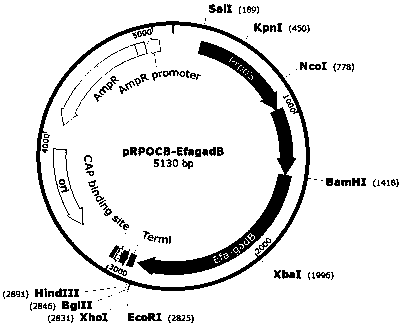
Image
Examples
Embodiment 1
[0080] The construction of embodiment 1 genetically engineered bacteria Escherichia coli (DH5α-LNSF1)
[0081] S1. Insert Enterococcus faecium LNSF2 into PSB or MRS medium, culture at 37°C for 12 hours, take 5mL of the culture solution and centrifuge at 8500r / min, collect the bacterial precipitate, discard the supernatant, and follow the Kangwei Century Bacterial Genome Extraction Kit Instructions Extract genomic DNA and store at -20°C for later use; design efa-1 and efa-2 primers, the biological sequences of primers efa-1 and efa-2 are shown in Table 1, and entrust Sangon Bioengineering (Shanghai) Co., Ltd. to synthesize ;Using efa-1 and efa-2 as primers, using Enterococcus faecium LNSF2 genomic DNA as a template, and using full gold TransStart Fast Pfu DNA polymerase for PCR reaction;
[0082]The PCR reaction conditions are: 95°C for 2min; 95°C for 20s, 50°C for 30s, 72°C for 55s, cycle 33 times; 72°C for 5min;
[0083] After the PCR reaction was completed, 1 μL Taq DNA Pol...
Embodiment 2
[0093] Example 2 Preparation of phosphoric acid modified chitin immobilized insoluble carrier
[0094] SS1. Add distilled water 3 times the mass of chitin, mix thoroughly to prepare chitin slurry, stir rapidly and slowly add concentrated phosphoric acid that is 50 times the mass of chitin that has been pre-frozen at -7°C under ice bath conditions, Stir until the mixture becomes transparent, ice bath for 1h (stir occasionally);
[0095] SS2. Under rapid stirring, add ice water 500 times the mass of chitin to the solution obtained in step SS1 in 4 times, let it stand for 30 minutes to fully precipitate the chitin, centrifuge at 4°C and 8500r / min for 15 minutes, and collect the chitin precipitate;
[0096] SS3. Add ice water 20 times the weight of the chitin precipitate, stir to resuspend, then centrifuge at 4°C, 8500r / min for 15min, collect the chitin precipitate, repeat twice to wash away phosphoric acid;
[0097] SS4. Then add distilled water according to 20 times of the mass...
Embodiment 3
[0100] Example 3 Preparation of NaOH / urea mixed solution modified chitin immobilized insoluble carrier
[0101] SS1. Add NaOH / urea mixed solution 50 times its mass to chitin, stir and mix well, freeze at -30°C for 4 hours, thaw at room temperature and stir quickly, then freeze at -30°C for 4 hours, thaw at room temperature and stir quickly, repeat Freeze and thaw until the chitin dissolves into a transparent state;
[0102] SS2. Add ice water with 500 times the mass of chitin to the solution obtained in step SS1 while stirring, let it stand for 30 minutes to fully precipitate chitin, centrifuge at 4°C and 8500r / min for 15 minutes, remove the supernatant, and collect the chitin precipitate;
[0103] SS3. Repeat washing the chitin precipitate 4 times with distilled water 20 times of the chitin precipitation to remove NaOH and urea;
[0104] SS4. Add 20 times of distilled water to the chitin precipitation, stir and resuspend, use 1mol / L HCl to adjust the pH close to neutral, neu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com