Halohydrin dehalogenase mutant for improving enantioselectivity and application thereof
A halohydrin dehalogenase, enantioselective technology, applied in the fields of genetic engineering and enzyme engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0113] Implementation of Site-Directed Saturation Mutagenesis
[0114] Recombinant plasmid pET28a-HHDH Ab Design appropriate mutation primers for the mutation template (see Patent Publication No. CN107881182A for the construction method).
[0115] The primers used for site-directed mutagenesis were:
[0116]
[0117] The PCR amplification system is: 10 μL of 5×PS Buffer, 4 μL of dNTP (2.5 mM each), 0.5 μL of forward and reverse mutation primers, 0.5 μL of template plasmid, 0.5 μL of PrimeSTAR DNA polymerase, and make up to 50 μL.
[0118] PCR conditions were pre-denaturation at 98°C for 2 minutes, 25 cycles: 98°C for 10s, 65°C for 10s, 72°C for 6min, and finally 72°C for 10min. Take 20 μL of PCR solution, add 1 μL Dpn I, digest at 37°C for 2-3 hours to remove plasmid DNA as a template, inactivate at 65°C for 10 minutes, immediately transform competent cells E.coli BL21(DE3), and coat with kanamycin (50mg / L) LB plates, cultivated at 37°C, picked positive transformants for...
Embodiment 2
[0121] Inducible expression of mutants and parental
[0122] The recombinant engineered strain prepared in Example 1 was cultured in 50 mL LB medium with a final mass concentration of 50 mg / L kanamycin at 37° C. and 200 r / min for 10 h. Then inoculate into the new 50mL LB culture medium that contains the kanamycin of 50mg / L with the inoculum size of 1vt.%, still with 37 ℃, 200r / min culture, treat to be cultivated to optical density (OD 600 ) is 0.6-0.8, add isopropyl-β-D-thiogalactopyranoside (IPTG) inducer to a final concentration of 0.15mM, and induce expression at 28°C and 200r / min for 12h. 5000 × g, 5min centrifugation to collect the bacterial cells, and NaH with pH 8.0 2 PO 4 -Na 2 HPO 4 The buffer was resuspended and washed, centrifuged at 5000×g for 5 min, and E. coli cells were collected and stored at -20°C for future use.
Embodiment 3
[0124] Determination of enantioselectivity of mutant enzymes
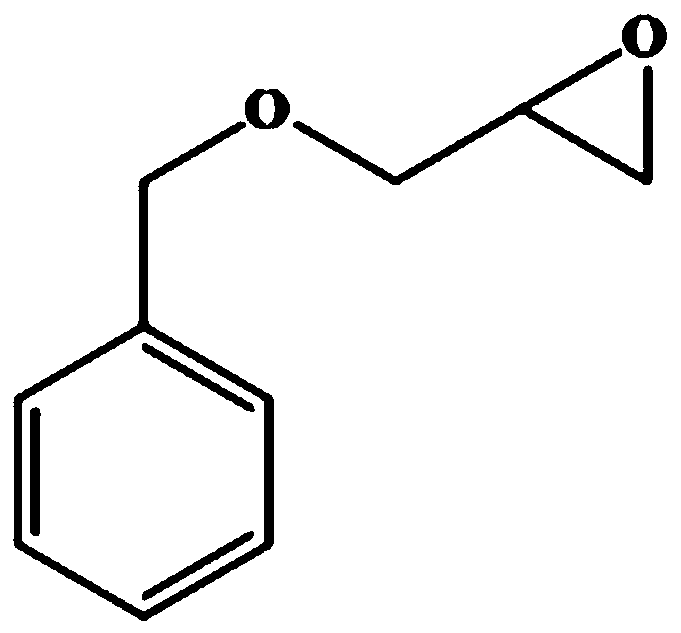
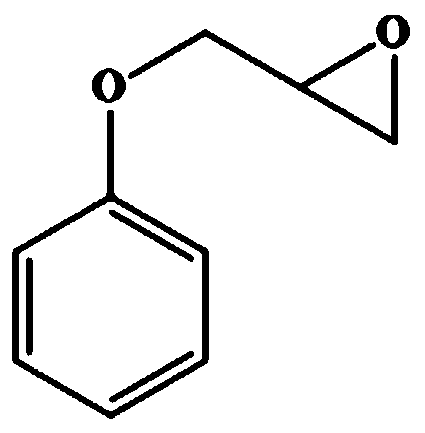
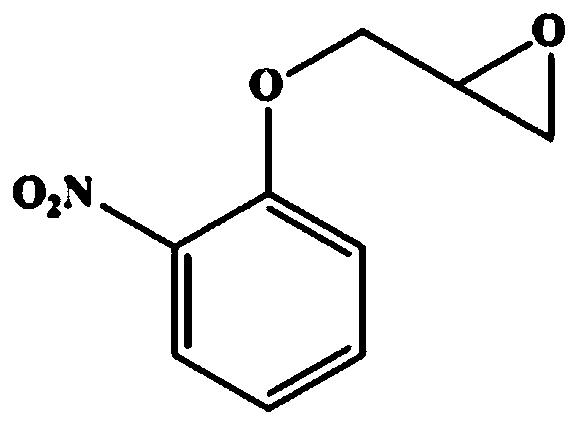
[0125] Substrate benzyl glycidyl ether (structural formula sees figure 1 ) HPLC analysis method: using Agilent-1220 system, column type: Chiralcel OD-H column (Daicel Co., Japan; 4.6×250mM, 5μm); chromatographic conditions: column temperature 30°C; mobile phase: n-hexane : Isopropanol=9:1 (v / v); Flow rate: 0.8mL / min; UV wavelength is 254nm; (S)- and (R)-substrate retention times (min) are about 9.3 and 10.1 respectively. Substrate phenyl glycidyl ether (structural formula sees figure 2 ) HPLC analysis method: using Agilent-1220 system, column type: Chiralcel OD-H column (Daicel Co., Japan; 4.6×250mM, 5μm); chromatographic conditions: column temperature 30°C; mobile phase: n-hexane : Virahol=83:17 (v / v); Flow rate: 0.8mL / min; UV wavelength is 220nm; (R)- and (S)-substrate retention times (min) are about 8.1 and 12.4 respectively. Substrate o-nitrophenyl glycidyl ether (structural formula sees image 3 ) HPLC an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com