Method for detectingprotein interaction by co-immunoprecipitation on basis of two-color fluorescent tag proteins GFP and mCherry
A technology of co-immunoprecipitation and labeling proteins, which is applied in material analysis by optical means, fluorescence/phosphorescence, measuring devices, etc. It can solve problems such as difficult real-time monitoring of target protein full expression, time-consuming, and difficult results.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] A method for detecting protein interaction by co-immunoprecipitation based on two-color fluorescent label protein GFP and mCherry, comprising the following steps:
[0056] (1) Construct vectors containing target genes to be verified for interaction
[0057] The In-Fusion cloning reaction system was established, and the two target genes to be verified for interaction were homologously recombined into the fluorescent transient expression vectors pBinplus-GFP (CN201711362357.4) and pBinplus-mCherry (Li et al., 2019), the plasmid was extracted and transformed into Agrobacterium tumefaciens GV3101 by electric shock (Li et al., 2019).
[0058] (2) Instantly transform tobacco leaves after mixing the above-mentioned Agrobacterium infection solution in equal proportions
[0059] Dip the above-mentioned GV3101 bacterial liquid with correct bacterial inspection into the 4~6ml LB liquid medium containing 50ug / ml kanamycin and 50ug / ml rifampicin resistance respectively, and shake c...
Embodiment 2
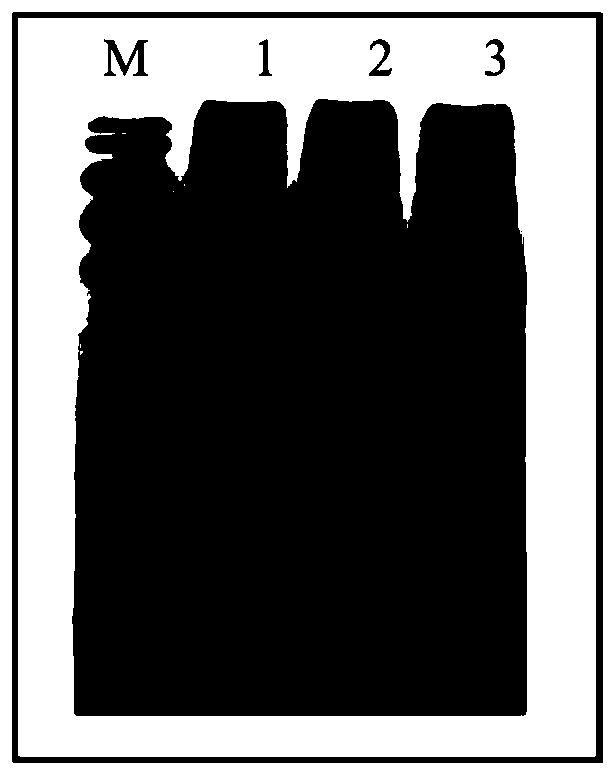
[0068] The exogenous Cry1Ab / c protein (Tu et al., 1998) expressed in the insect-resistant transgenic rice Huahui No. 1 (Tu et al., 1998) and three kinds of endogenous rice protein calmodulin binding transcription factor CAMTAs of different molecular weights were used in the specific method of Example 1 (Gene ID: XM_015758912.2), 3-deoxy-D-arabinose heptulose-7-phosphate DAHP (Gene ID: XM_015790892.1), histone lysine methyltransferase HKMTs (Gene ID : XM_015792076.2) to detect protein interaction by co-immunoprecipitation, the above three proteins are rice endogenous proteins that can interact with exogenous Cry1Ab / c protein in yeast two-hybrid primary screening.
[0069] The specific construction method is as follows: refer to the method in Example 1 to establish the In-Fusion cloning reaction system, homologously recombine the cry1Ab / c gene into the N-terminus of the fluorescent transient expression vector pBinplus-GFP through the endonuclease BamH, and respectively integrate ...
Embodiment 3
[0076] Example 3 Verify the validity of the above co-immunoprecipitation results by bimolecular fluorescence complementary technology
[0077] According to the method of Example 1, the In-Fusion cloning reaction system was established, and the cry1Ab / c gene was homologously recombined into the C-terminus of the bimolecular fluorescence complementary expression vector PCV-cYFP (Hu et al., 2015) by endonuclease BamH , homologous recombination of CAMTAs, DAHP and HKMTs genes into the C-terminus of the expression vector PCV-nYFP (Hu et al., 2015). The homologous recombination primers are shown in Table 3. After extracting the plasmid, electric shock transformed Agrobacterium GV3101 to obtain exogenous cry1Ab GV3101 infection liquid containing c-cYFP gene, GV3101 infection liquid containing CAMTAs-nYFP gene, GV3101 infection liquid containing DAHP-nYFP gene and GV3101 infection liquid containing HKMTs-nYFP gene.
[0078] The GV3101 infection solution containing the exogenous cry1Ab...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com