Method for detecting Fusobacterium nucleatum by using PCR-ELISA
A fusobacterium nucleatum, purpose technology, applied in the field of molecular biology detection, can solve the problems of low sensitivity, difficult to master, low cost, etc., and achieve good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The PCR amplification of embodiment 1FadA gene
[0029] According to the FadA gene of F. nucleatum in GeneBank, a pair of specific primers were designed and synthesized by Shanghai Bioengineering Co., Ltd.
[0030] The primer sequences are as follows:
[0031] F: 5'-CACAAGCTGACGCTGCTAGA-3'; SEQ ID NO.1;
[0032] R: 5'-TTACCAGCTCTTAAAGCTTG-3'; SEQ ID NO.2.
[0033] The freeze-dried powder of F. nucleatum was activated, operated in an anaerobic box with tryptone soybean liquid medium, the bacterial liquid was collected, the genomic DNA of F. nucleatum was extracted, and the FadA gene was amplified by PCR using it as a template.
[0034] Wherein, the PCR reaction system is: template 2 μl, upstream primer 0.5 μl, downstream primer 0.5 μl, Premix Taq 25 μl, double distilled water 22 μl.
[0035] The PCR reaction conditions were: 94°C for 5min; 30 cycles of 94°C for 30s, 59°C for 30s, and 72°C for 30s; 72°C for 10min; and storage at 4°C.
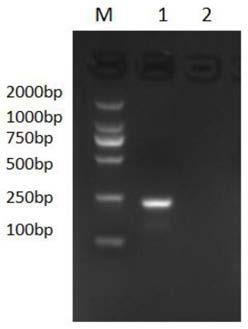
[0036] Use 1.5% agarose gel to c...
Embodiment 2
[0037] The construction of embodiment 2 recombinant plasmids
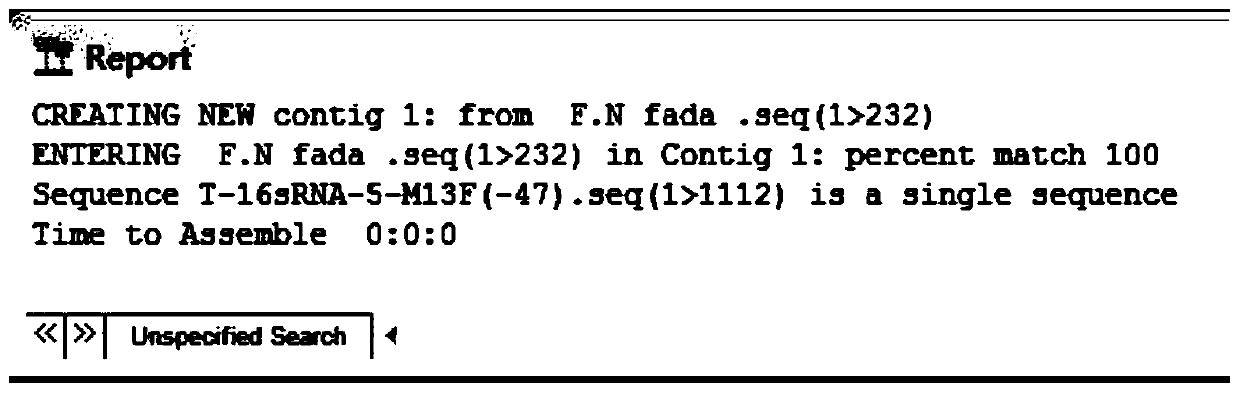
[0038] The amplified target gene was connected to the PMD-18-T vector, and transformed into DH5α cells to construct a recombinant plasmid; the constructed recombinant plasmid was sent to Shanghai Bioengineering Co., Ltd. for sequencing, and the whole genome sequence of Fusobacterium nucleatum FadA was uploaded from NCBI. And compared with DNAman sequence analysis software, the result showed that the identity was 100% ( figure 2 ), consistent with the expected results, the target fragment size is 232bp.
Embodiment 3
[0039] Embodiment 3PCR-ELISA detects
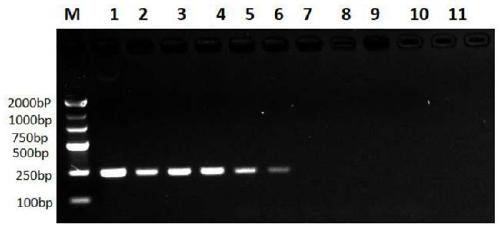
[0040](1) The primers in Example 1 are labeled, wherein the 5' end of the upstream primer F is labeled with digoxin, and the 5' end of the downstream primer R is labeled with biotin; the recombinant plasmid constructed in Example 2 is used as a template, according to The conditions of Example 1 were used for PCR amplification, and then for ELISA detection.
[0041] (2) Dilute the streptavidin stock solution (1mg / ml) with carbonate buffer solution with pH 9.5, and add 100μl per well to the microtiter plate to coat, each sample is repeated three times, overnight at 4°C ; Wash 3 times with PBST, 3 min each time; Block with 2% BSA, 200 μl per well, place at 37°C for 2 h, wash the plate 3 times as well; Dilute the amplified product using the recombinant plasmid as template with PBST, 100 μl well, place at 37°C for 1 hour, wash the plate 4 times, 3 minutes each time; dilute horseradish peroxidase-labeled anti-digoxigenin antibody with PBST, 10...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com