SERS-SPR dual-mode sensor as well as preparation method and application thereof
A sensor and detection method technology, applied in the field of functional nanomaterials and biological detection, can solve problems such as insufficient sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1 Preparation of silver nanohole-nanorod array substrate
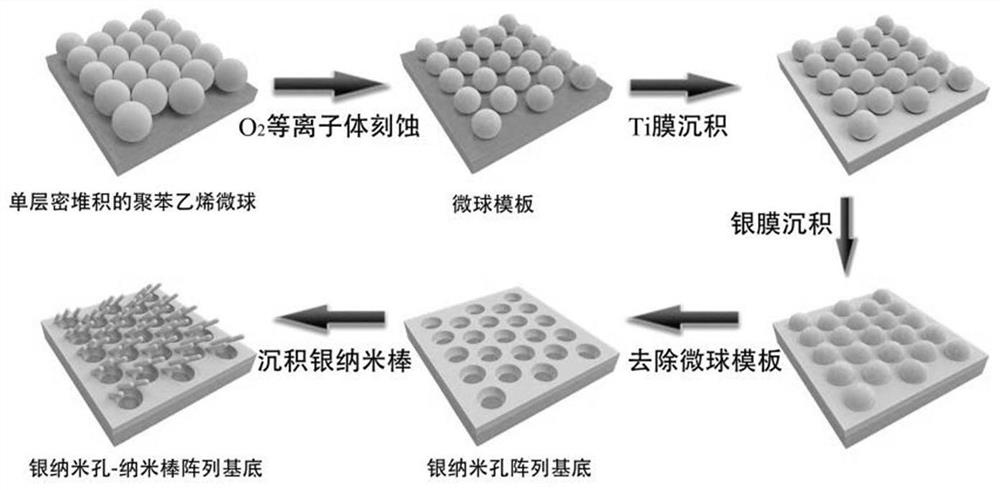
[0063] 1. Silver nanohole-nanorod arrays were prepared by nanosphere etching (NSL), reactive ion etching (RIE) and physical evaporation deposition techniques. figure 1 The fabrication process of the silver nanopore-nanorod array is shown. In the first step, 500nm polystyrene (PS) microspheres are assembled on a clean glass or silicon wafer in a monolayer close-packed manner by the air-water interface method to form a microsphere template, a glass or silicon wafer The size is 0.9×2.5em.
[0064] 2. In the second step, use O 2The plasma etches the PS microspheres on the surface of the substrate to reduce the diameter of the microspheres on the surface of the substrate. In the etching process, Trion Technology Phantom III RIE / ICP system was used to operate, the pressure was 40mTorr, the oxygen flow rate was 10sccm, the ICP power was 25W, the RF power was 10W, and the duration was 350s.
[0065] 3. Ph...
Embodiment 2
[0067] Example 2 Characterization of Refractive Index Response Performance of Silver Nanohole-Nanorod Array Substrate
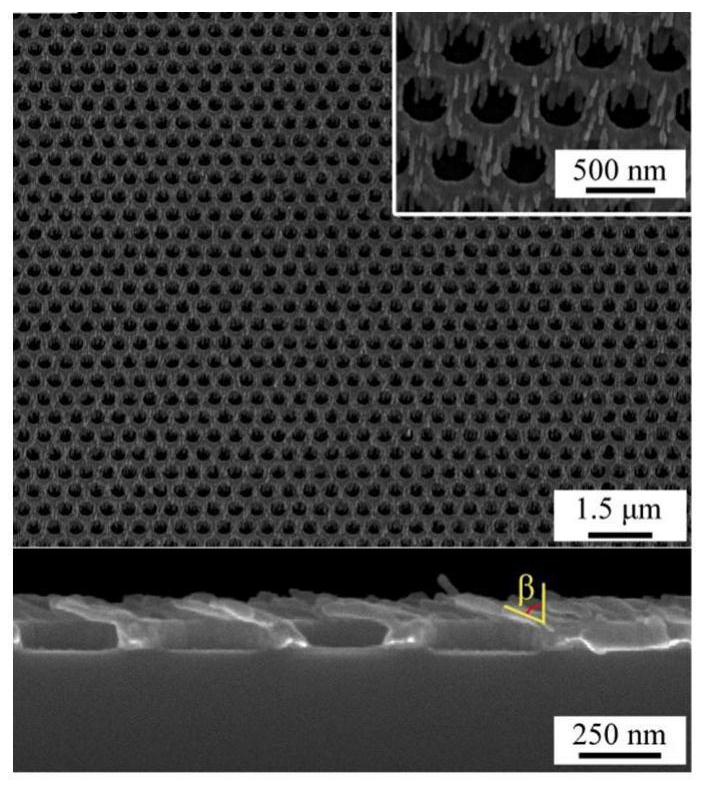
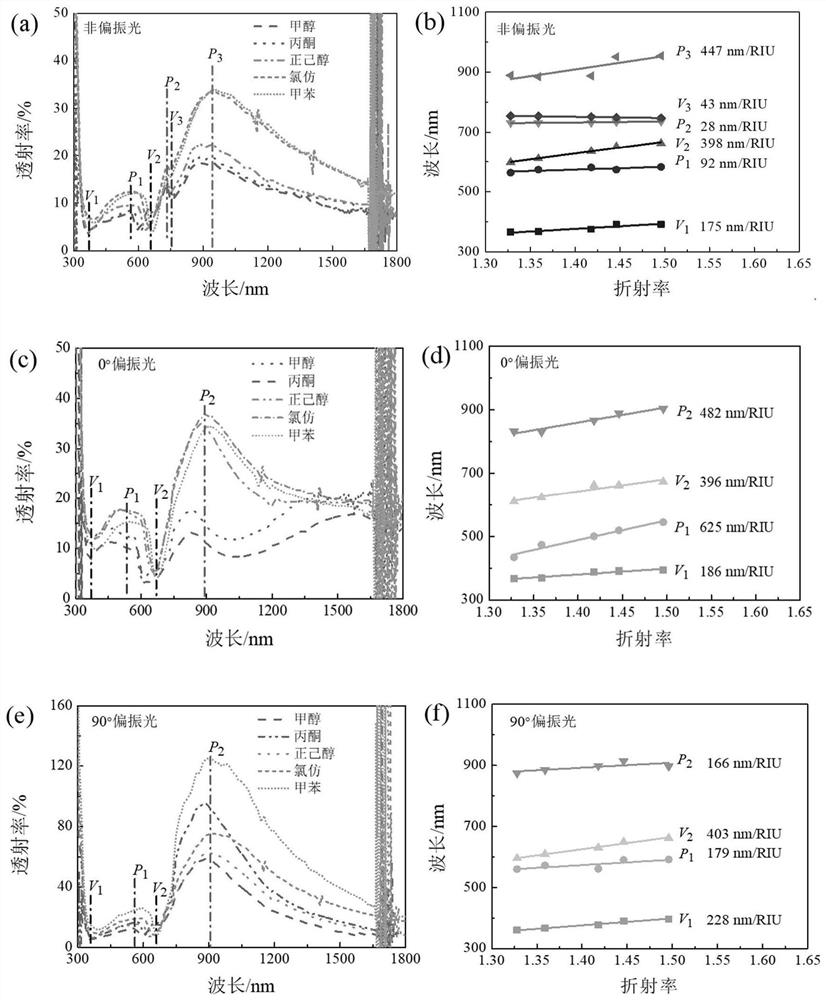
[0068] The silver nanohole-nanorod array substrate (structure such as figure 2 shown) were soaked in different refractive index solvents: methanol (n=1.328), acetone (n=1.359), 1-hexanol (n=1.418), chloroform (n=1.446) and toluene (n=1.496), Afterwards, the polarized and non-polarized transmission spectra of the silver nanohole-nanorod array substrate were tested respectively. Under the irradiation of non-polarized, 0-degree polarized and 90-degree polarized light, the transmission spectrum of the silver nanohole-nanorod array substrate exhibits a refractive index dependence, such as image 3 shown. In addition, both the spectral profile and the peak-to-valley variation of the transmission spectrum exhibit obvious polarization dependence. According to the response of different peaks and valleys to different refractive index solvents, the response of peaks...
Embodiment 3
[0069] Example 3 Preparation of SERS-SPR dual-mode sensor
[0070] 1. Preparation of tetrahedral DNA probes. Such as Figure 5 As shown, tetrahedral DNA is formed by base complementary hybridization self-assembly of four single strands A, B, C, and D. Mix equimolar amounts (1 μM) of four DNA single strands in 300 μL TM buffer (20 mM Tris-HCl, 50 mM MgCl2, pH 8.0), mix them, and anneal (heat to 95 ° C for 5 min, then naturally cool to room temperature) ), forming tetrahedral DNA, the concentration of tetrahedral DNA in the final solution was 1 μM. Tetrahedral DNA formation was characterized by 10% polyacrylamide gel electrophoresis, Figure 6 a is the electrophoretic gel image corresponding to the formation process of tetrahedral DNA. Compared with the combination of one (lane 1), two (lane 2) and three DNA strands (lane 3), four DNA strands are mixed and self-assembled to form a tetrahedron DNA moved the slowest in lane 4, indicating that tetrahedral DNA was formed success...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com