Three points three p450 enzyme and its application in the preparation of tropinone
A technology of tropinone and reductase, which is applied in the fields of biosynthesis and enzyme catalysis, and can solve the problems of large environmental pollution by chemical methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1 3
[0071] Example 1 Cloning of three-thirds cytochrome P450 and cytochrome P450 reductase genes
[0072] 1.1 Extraction and detection of total RNA
[0073] Turn on the ultra-clean bench and ultraviolet lamp, burn the mortar, keys, and scissors with ethanol. Cut off 100mg of three-thirds hairy roots, grind the tissue into powder in liquid nitrogen, and distribute it into Ep tubes. After the liquid nitrogen evaporates, add 1ml Trizol-x-100, shake vigorously immediately or use a pipette to blow 5- 8 times (until there are no lumps), let stand at room temperature for 5 minutes; extract twice with an equal volume of chloroform, and centrifuge at 7500g for 15 minutes; Centrifuge at 10,000g for 10min at ℃; add 1ml of 75% ethanol to the precipitate for washing, and centrifuge at 10,000g at 4°C for 10min; dry the precipitate at room temperature for 10min and dissolve in 25μL of DEPC-treated water, and use 1.0% agarose gel electrophoresis to detect the integrity of the RNA. The ratio and...
Embodiment 2
[0083] The construction of embodiment 2 eukaryotic expression vector
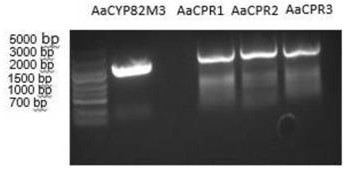
[0084] 2.1 Cloning of AaCYP82M3 and AaCPR1-3 genes: Use Axygen’s plasmid extraction kit to extract and sequence the correct T vector, use the AaCYP82M3 and AaCPR1-3 genes in the T vector as templates, and use the primers in Table 1 to perform PCR amplification. The following system [5×phusion buffer 4.0μL, dNTP (2.5mM) 1.6μL, each primer (2μM) 2μL, DMSO 0.6μL, cDNA 0.5μL, Phusion DNA polymerase 0.2μL, add ddH 2 O to a final volume of 20.0 μL] to amplify the gene, the reaction program: pre-denaturation at 98°C for 30s, denaturation at 98°C for 10s, annealing at 55°C for 30s, extension at 72°C for 2min, 30 cycles, final extension at 72°C for 5min, gel recovery using Axygen Reagents are recovered to obtain the corresponding fragments.
[0085] 2.2 Gene double digestion: Digest the PCR product of AaCYP82M3 with restriction enzymes BamHI and KpnI at 37°C for 2h; digest the PCR product of AaCPR1 with restriction...
Embodiment 3
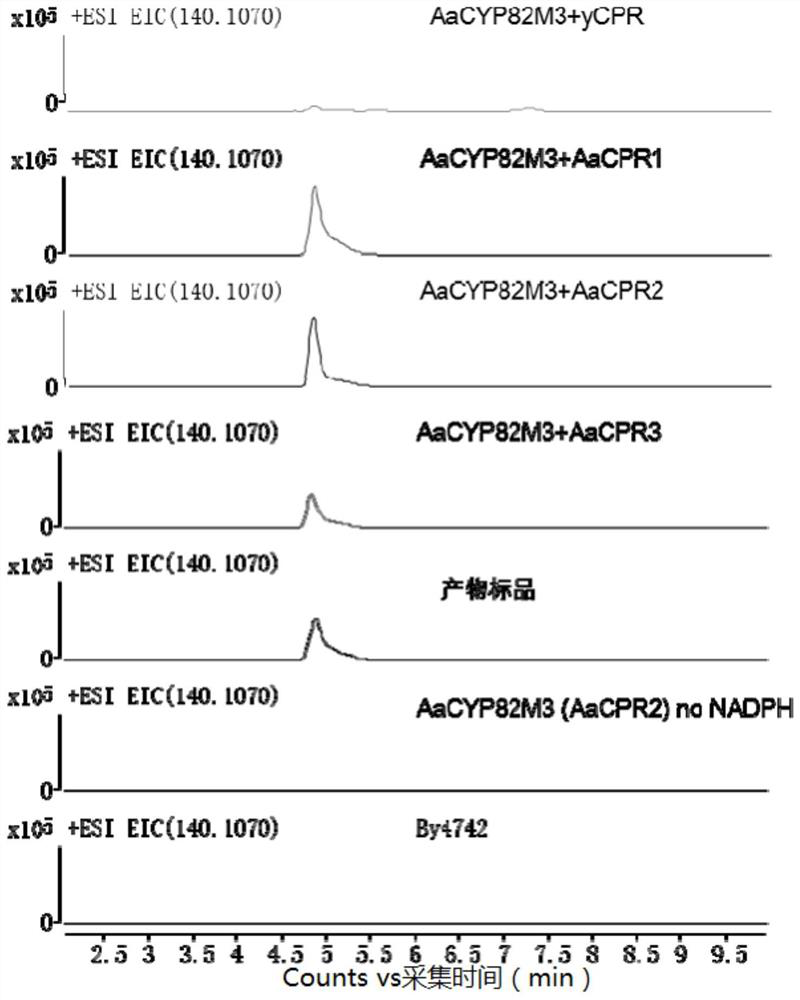
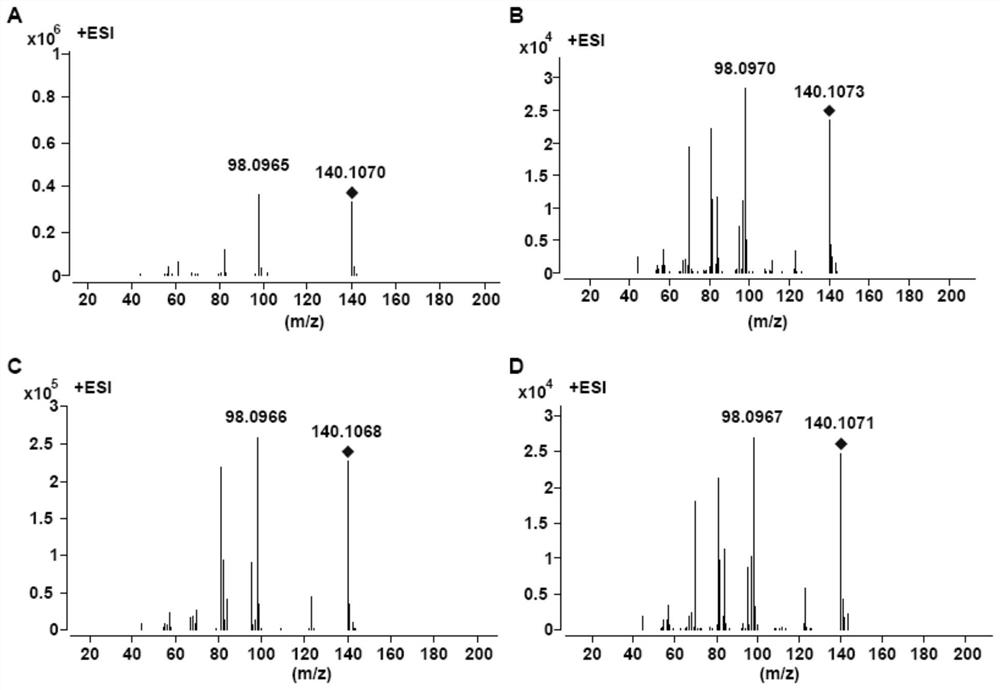
[0089] Example 3 Construction of AaCYP82M3 and AaCPR1-3 Expression Strains
[0090] Saccharomyces cerevisiae By4742 was streaked on a YPD plate and cultured at 30°C for 2 days; a single colony of yeast was picked and placed in 5mL YPAD liquid medium, cultured overnight at 30°C and 200rpm; the yeast liquid was diluted 10 times, and the OD was measured 600 , at this time OD 600 Should be 0.4-0.5 (indicating good growth). Transfer to 2*YPAD (fast growth) at a ratio of 1:20 to 1:25, and culture in 50mL to make the initial OD 600 Less than or equal to 0.2; cultivate to OD at 30°C, 200rpm 600 0.5, about 4-5 hours; the carrier DNA (protist DNA) frozen at -20°C was denatured at 100°C for 5 minutes, and immediately inserted into the ice (keep single strand). Centrifuge at 1,000g for 5 minutes at room temperature to collect bacteria, resuspend with 1 / 2 volume of sterile water (for example, resuspend 50mL of bacterial liquid with 25mL of sterile water) and centrifuge; wash again with ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com