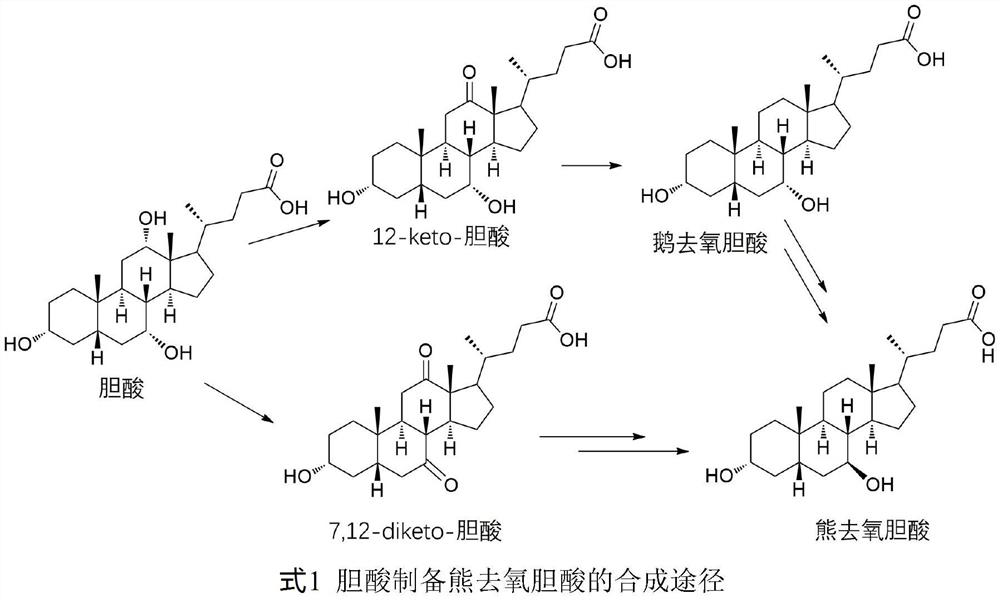
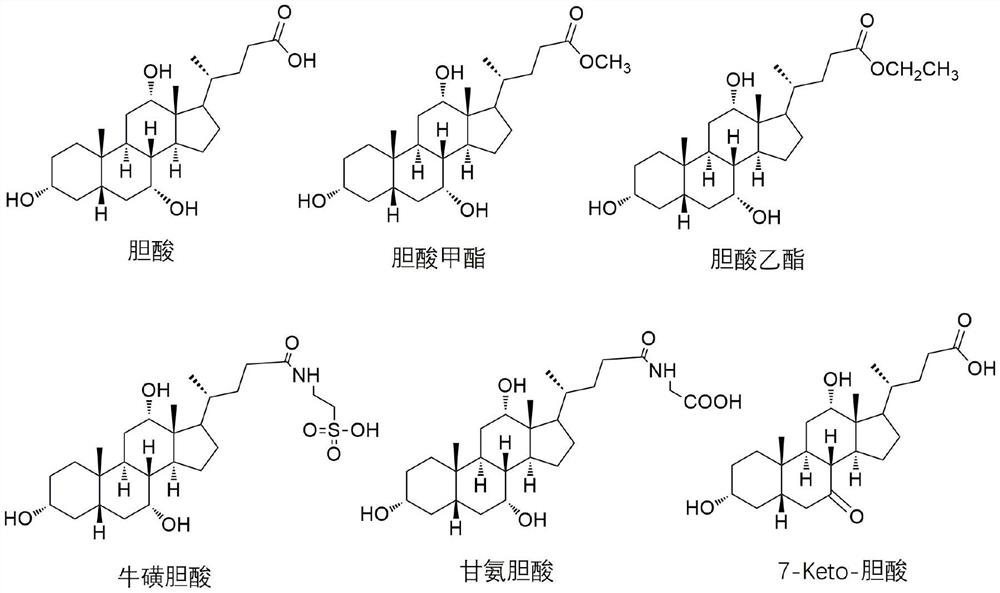
12-hydroxycholic acid dehydrogenase and application thereof
A technology of hydroxycholic acid dehydrogenase and hydroxycholic acid, applied in the direction of oxidoreductase, microorganism-based methods, microorganisms, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0013] Example 1: Synthesis of 12-hydroxycholic acid dehydrogenase gene and construction of genetically engineered bacteria
[0014] 1.1 Synthesis of 12-hydroxycholic acid dehydrogenase gene
[0015] The possible 12-hydroxycholic acid dehydrogenase sequence from Eggerthella lenta was mined by gene mining method, and the gene was synthesized by codon optimization according to the protein sequence, and constructed into pET21a expression vector, the gene insertion sites were NdeI and XhoI.
[0016] 1.2 Transformation of recombinant plasmids
[0017] Competent Escherichia coli cells were prepared by calcium chloride method.
[0018] (1) Put 10 μL of the recombinant plasmid in 50 μL of Escherichia coli BL21(DE3) competent cells, and place in ice bath for 30 minutes.
[0019] (2) Heat-shock in a water bath at 42°C for 45 seconds, and quickly place on ice for 1-2 minutes.
[0020] (3) Add 600 μL of fresh LB liquid medium, shake and culture at 37°C for 45-60min.
[0021] (4) Take ...
Embodiment 2
[0022] Embodiment 2: the construction of co-expression bacteria
[0023] Recover the pRSFDuet-flavin reductase vector already in the laboratory with NdeI / XhoI double enzyme digestion, and recover the gene fragment after pET-12-steroid dehydrogenase double enzyme digestion with NdeI / XhoI, and use T4 DNA for the fragment and linear vector After ligation with ligase, BL21(DE3) was transformed into competent cells, single clones were picked for verification and activity detection, and pRSFDuet-flavin reductase-12-hydroxysteroid dehydrogenase co-expression strains were obtained.
[0024] Recover the pRSFDuet-lactate dehydrogenase vector already in the laboratory with NdeI / XhoI double enzyme digestion, pET-12-steroid dehydrogenase with NdeI / XhoI double enzyme digestion and recover gene fragments, and T4 DNA for fragments and linear vectors After ligase ligation, BL21(DE3) was transformed into competent cells, single clones were picked for verification and activity detection, and pRS...
Embodiment 3
[0025] Example 3: Induced expression of single-expression bacteria and co-expression bacteria
[0026] Prepare 50 mL of seed liquid, and the medium is LB liquid medium (peptone 10g / L, yeast powder 5g / L, NaCl 10g / L), pick a single colony of genetically engineered bacteria with an inoculation loop and insert it into the medium, 37°C, 200rpm Incubate overnight. The overnight cultured seed solution was transferred to the fermentation medium (industrial medium) at an inoculum size of 1%, and cultured at 32° C. and 200 rpm for 20 h. After 1 mL of the fermentation liquid was ultrasonically disrupted, the activities of 12-hydroxycholic acid dehydrogenase and flavin reductase were detected. The definition of 12-hydroxycholic acid dehydrogenase enzyme activity: the amount of enzyme required to generate 1 μmol NADH per minute is 1 enzyme activity unit (U); the definition of enzyme activity of flavin: the amount of enzyme required to consume 1 μmol NADH per minute is 1 enzyme activity u...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com