Novel efficient beta-glucosidase CsBGL as well as encoding gene and application thereof
A technology of glucosidase and coding gene, applied in the field of β-glucosidase CsBGL and its coding gene and application, and biology, can solve the problems of low efficiency, low enzyme yield, poor specificity, etc., and achieve operational production The method is simple, the catalytic specificity is strong, and the effect of good industrialization prospect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
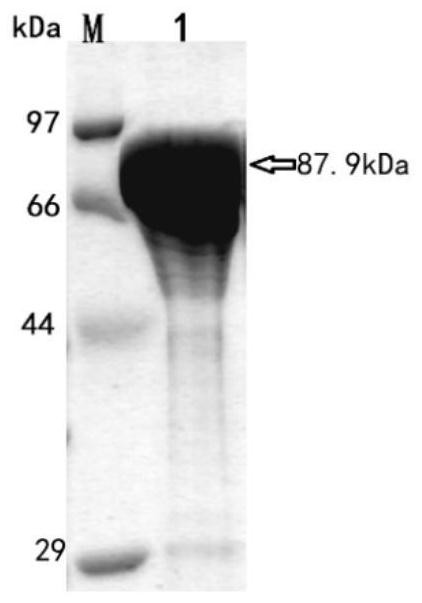
[0045] Embodiment 1: Preparation of the crude enzyme liquid of β-glucosidase CsBGL
[0046] Chryseobacterium sp. 1433 was inoculated in LB antibiotic-free medium, cultured at 30°C and 200rpm for 24h, and then centrifuged at 3000-10000rpm for 2-10min to collect bacterial precipitates. With 50mM Na at pH 6.0-8.0 2 HPO 4 / NaH 2 PO 4 Resuspend the cells in the buffer, then centrifuge at 3000-10000rpm for 2-10min to collect the bacterial pellet again, and then use 10-20mL of 50mM Na at pH 6.0-8.0 2 HPO 4 / NaH 2 PO 4 The cells were resuspended in buffer, disrupted by ultrasonic waves, centrifuged at 12,000 rpm for 20 min, and the supernatant was collected as crude enzyme solution of β-glucosidase CsBGL.
Embodiment 2
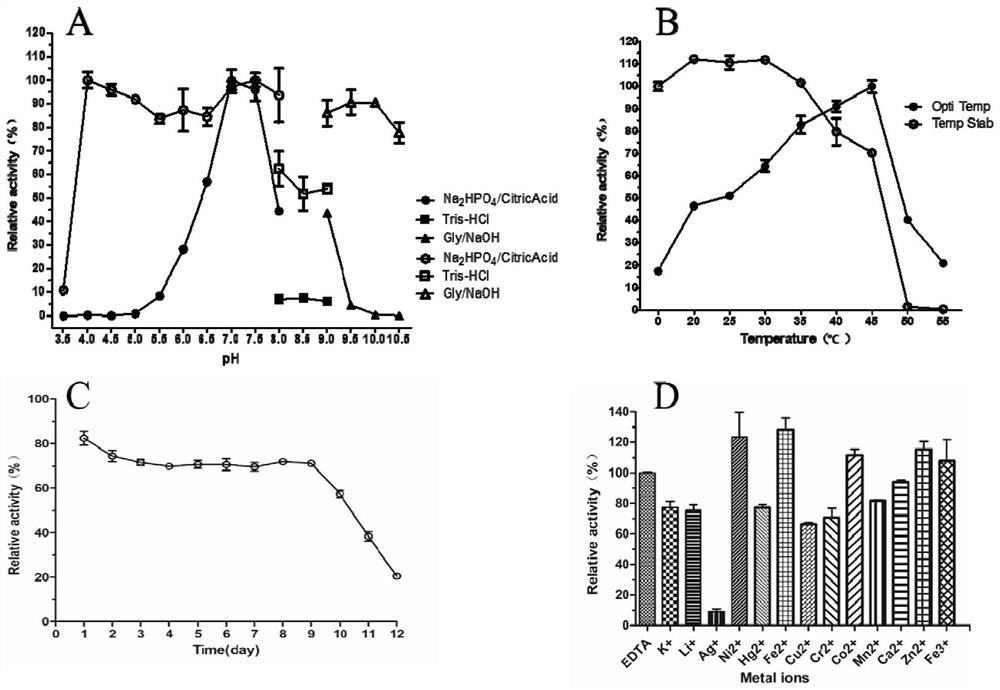
[0047] Embodiment 2: Native-PAGE electrophoresis under acidic conditions of β-glucosidase CsBGL crude enzyme solution
[0048] The β-glucosidase CsBGL crude enzyme solution obtained in Example 1 was subjected to Native-PAGE electrophoresis. The specific parameters of the electrophoresis were: the positive and negative electrodes were inverted, traced with methyl green (0.002%), and a constant current of 10mA was used at 4°C. Electrophoresis 2 ~ 3h.
[0049] The formula of Native-PAGE gel under acidic conditions is as follows:
[0050] 1) Separating gel: 0.06M KOH, 0.376M HAc, pH 4.3 (7.7% gel concentration, acrylamide:methylene acrylamide is 37.5:1);
[0051] 2) Stacking gel: 0.06M KOH, 0.063M HAc, pH 6.8 (3.125% gel concentration, acrylamide: methylene acrylamide is 3:1);
[0052] 3) Electrophoresis buffer: 0.14M L-alanine, 0.35M glacial acetic acid, pH 4.5.
[0053] Use a clean scalpel to cut off the corresponding swimming lane in the PAGE gel, and use pH7.4, 50mM Na 2 H...
Embodiment 3
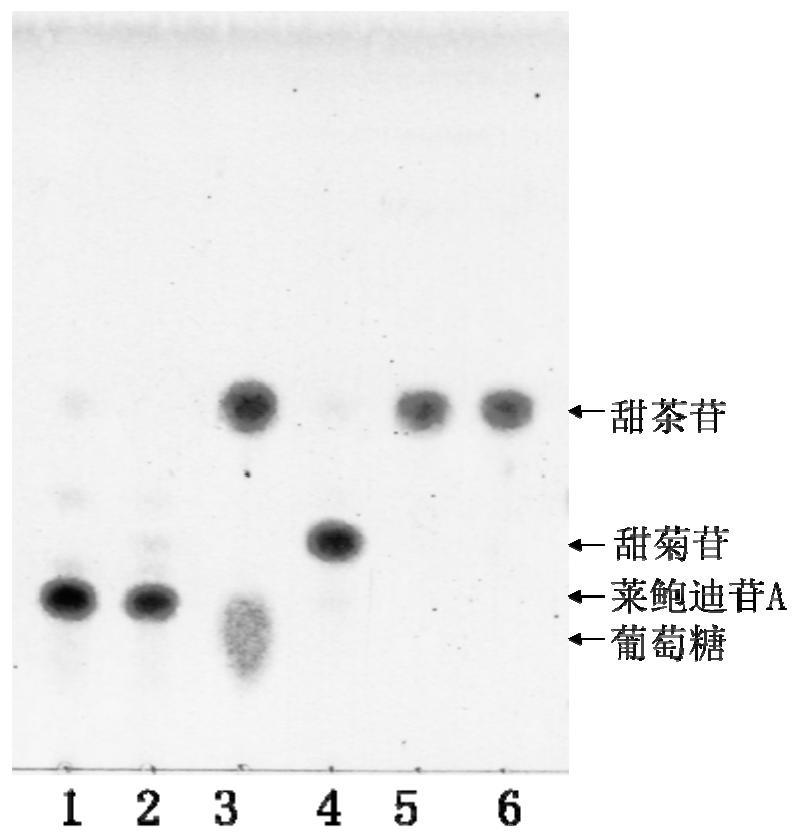
[0054] Embodiment 3: Cloning of gene encoding β-glucosidase CsBGL, construction of recombinant vector and heterologous expression
[0055] 1. Extraction of Genomic DNA from Chryseobacterium sp. 1433
[0056] Chryseobacterium sp. 1433 was inoculated in LB medium, cultured at 30° C. and 200 rpm for 24 hours, and then centrifuged at 3000 to 10000 rpm for 2 to 10 minutes to collect bacterial precipitates. Using the Tiangen Genomic DNA Extraction Kit, follow the steps in the manual to extract DNA from the collected bacterial precipitates to obtain Chryseobacterium sp. 1433 genomic DNA.
[0057] 2. Identification of the amino acid sequence of β-glucosidase CsBGL and the nucleotide sequence of the coding gene
[0058] The genomic DNA of Chryseobacterium sp. 1433 extracted in step 1 was sequenced.
[0059] The Native-PAGE electrophoresis activity staining band that embodiment 2 obtains is cut into 1mm 3 Size, in-gel digestion with trypsin, and comparison with the Chryseobacterium s...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com