R-loop binding protein GST-His6-1/2*HBD and whole genome R-loop detection method
A technology that combines proteins and detection methods, applied in the field of nucleic acid omics, can solve the problems of cumbersome operation steps, poor signal background, and long experiment cycle, and achieve the effects of improved detection efficiency, high signal-to-noise ratio, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
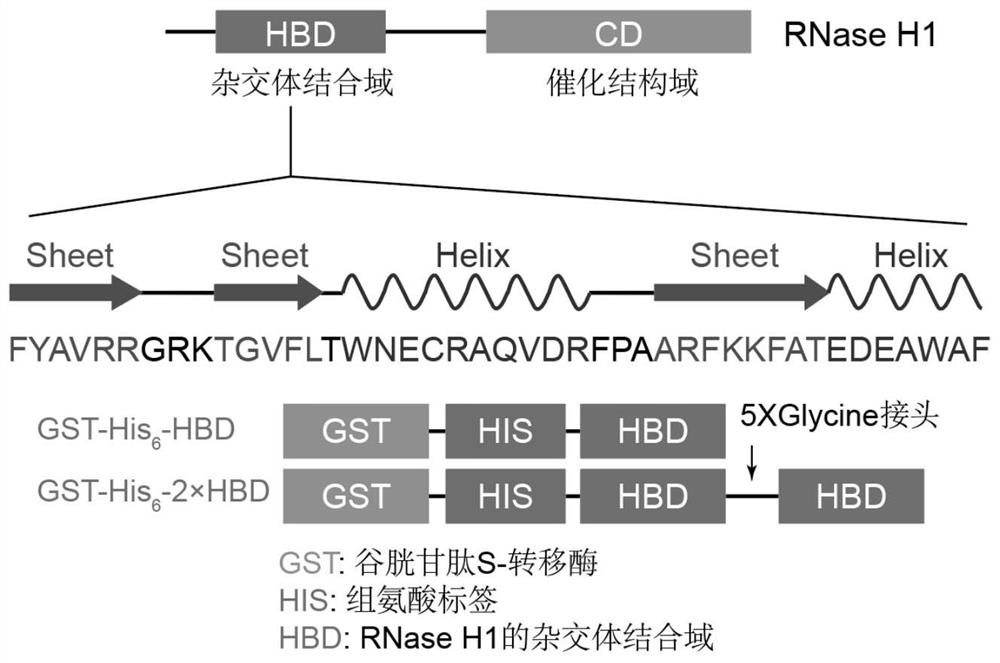
[0039] [Example 1] Recombinant plasmid GST-His 6 -1 / 2 x HBD build
[0040] 1. Design and synthesis of primers
[0041] According to the high homology of RNase H1 among different species provided by Marcin Nowotny et al. (2008), the ppyCAG-RNASEH1-D210N plasmid (addgene#111904) containing human RNase H1 coding sequence was purchased from addgene, and designed according to the plasmid information Synthetic specific primers amplify the HBD domain. Five glycine repeat sequences were added to one primer to express a flexible linker to promote tandem HBD protein expression. The primers were synthesized by Shanghai Sangon Bioengineering Technology Co., Ltd., and the sequences and numbers of each primer are as follows:
[0042] (1) HBD gene amplification primers
[0043] LP-1:
[0044] GCCATATCGAAGGTCGTCATATGATGTTCTATGCCGTGAGGAGGGGC
[0045] RP-1:
[0046] CTTTGTTAGCAGCCGGTTATCCCCCTCCGCCTCCGCTTGCAGATTTCCT
[0047] GACAAAGGC
[0048] (2) 2×HBD gene amplification primers
[00...
Embodiment 2
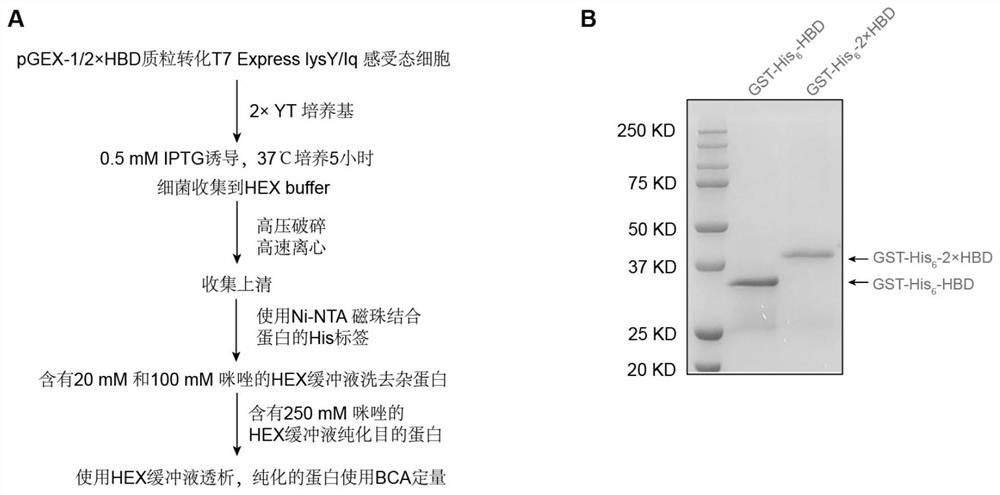
[0055] [Example 2] GST-His 6 -1 / 2×HBD protein expression and purification
[0056] 1.GST-His 6 -1 / 2 x HBD protein expression
[0057] GST-His 6 -1 / 2×HBD recombinant plasmid transformed into T7 Express lysY / I q Competent cells, after resistance screening, selected monoclonal strains were cultured in 2×YT medium containing 100 μg / mL ampicillin sodium, and the bacterial culture conditions were set at 37°C and the rotation speed was 200rpm. when the bacterial OD 600 When 0.6 was reached, protein expression was induced by adding 0.5 mM IPTG. Bacteria particles were collected by centrifugation after cultivating for 5 hours, excess culture medium was removed, and lysis HEX buffer solution (components including 20mM HEPES, 0.8M sodium chloride, 10% glycerol, 0.2% Triton X-100, 1X protease inhibition) was added at pH 7.5 agent). Mix the buffer and the bacteria thoroughly, then use a cell disruptor to perform high-pressure disruption, break at 4°C and 800psi for 5 minutes, and th...
Embodiment 3
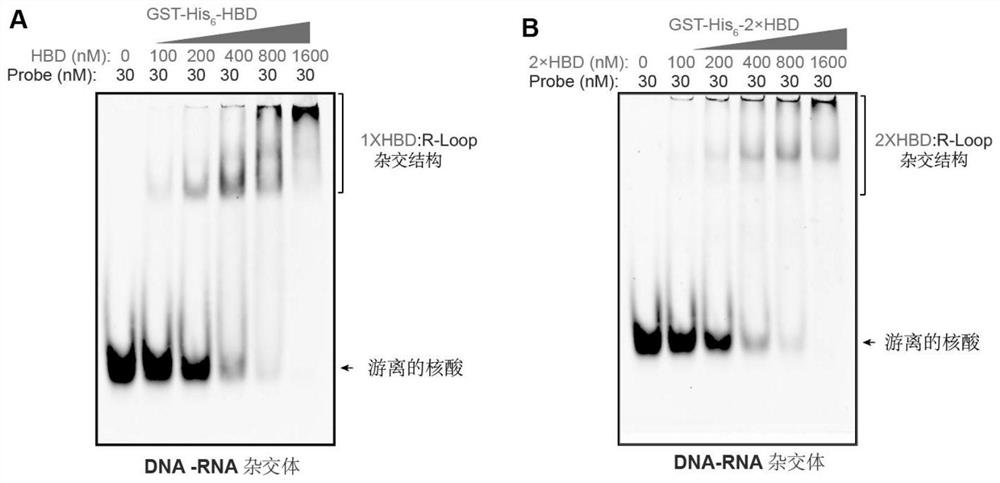
[0060] [Example 3] GST-His 6 -1 / 2×HBD protein specifically recognizes the R-loop structure
[0061] (1) Gel electrophoresis mobility assay (EMSA) to detect different concentrations of GST-His 6 The binding ability of -1 / 2×HBD protein and DNA:RNA hybrid, the reaction conditions and system are as follows:
[0062]
[0063] Mix the above components with a pipette, and perform the following reaction on a PCR thermal cycler: incubate at 25°C for 30 minutes
[0064] (2) Add the sample in step (1) to 6% TBE-PAGE gel for electrophoresis, the electrophoresis buffer is 0.5×TBE, the electrophoresis voltage is set to 60V, and the electrophoresis time is 60 minutes.
[0065] (3) Use GE's Typhoon9500 to scan the gel, and verify GST-His according to the blocking phenomenon of nucleic acid in the gel 6 -1 / 2×HBD protein can specifically bind to DNA:RNA hybrids, see the specific results image 3 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com