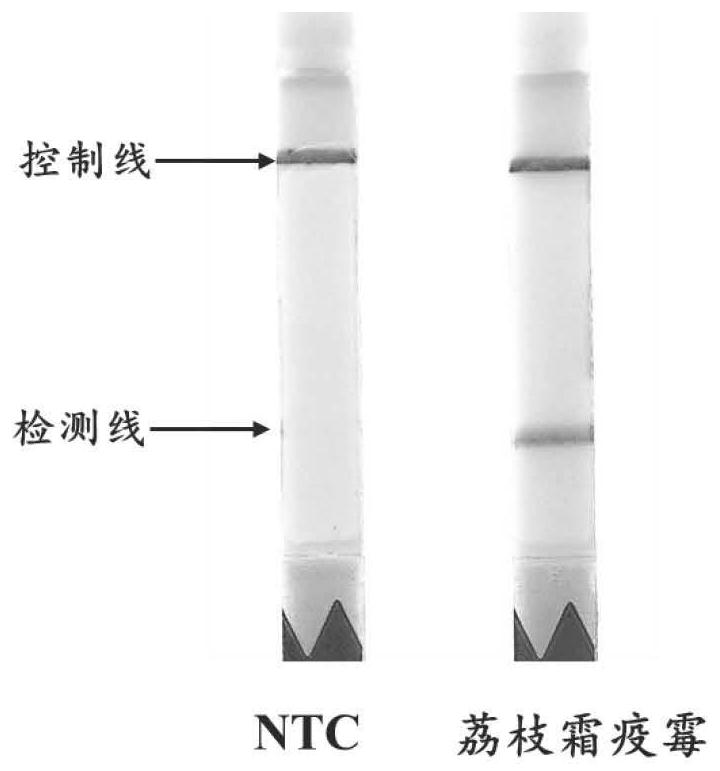
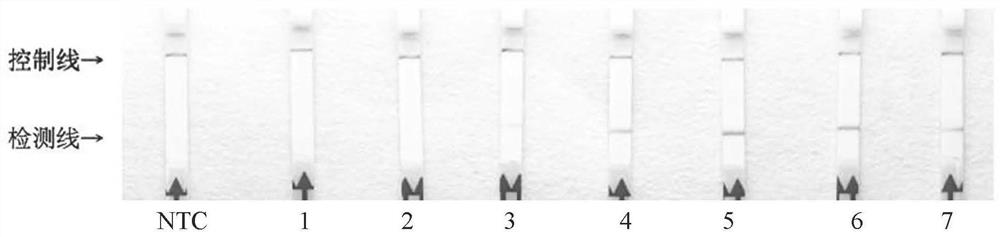
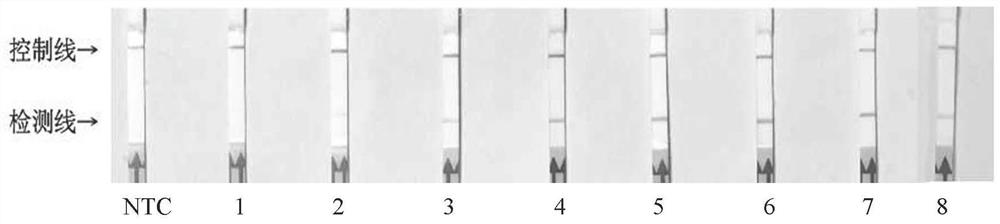
Primer and probe combination for detecting peronophythora litchii based on RPA-lateral flow chromatography technology and detection method of peronophythora litchii
A litchi downy mildew and lateral flow chromatography technology, applied in the biological field, can solve the problems of few plant pathogenic oomycetes and no related application reports, and achieve reliable results, simple and fast operation, and good practicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Design of primers and probes for the detection of Phytophthora lychee RPA-lateral flow chromatography
[0044] (1) Acquisition of Rh gene from Peronophythora litchi
[0045] The genome data of Pe.litchi was downloaded from GenBank (ID: PRJNA290406), and the hidden Markor of the ammonium transporter domain was downloaded from the Pfam database (http: / / pfam.sanger.ac.uk / ). Hidden Markov Model (HMM), model accession number is PF00909. The HMMER 3.2.1 program (http: / / hmmer.org / ) was used to search and compare the proteins in the whole genome of Phytophthora litchie, and the E-value threshold of the target functional domain was set to 1e-10. Subsequently, the compared candidate genes were submitted to NCBI for comparison and verification with the identified sequences, and the nucleotide sequence of the Rh gene was obtained, as shown in SEQ ID NO.1.
[0046] 5’-ATGTGTCTCATCTTCTTCGCCCTGAAATTCGACATGCCCAGTCCGAAAAACAACGACGAAGACACTGTCAGCACTCTAAACTACTACCCCATGTACATGGATGT...
Embodiment 2
[0054] Example 2 RPA-Lateral Flow Chromatography-based Detection Primer and Probe Combination for Visual Detection of Lychee Peronospora
[0055] (1) Extraction of Phytophthora lychee Genomic DNA:
[0056] Adopt CTAB method to extract Phytophthora lychee Genomic DNA, the specific process is as follows: get 50mg freeze-dried mycelia powder in a 1.5ml centrifuge tube, add 900 μ l 2% CTAB (cetyltrimethylammonium bromide) extract (2% CTAB; 100mmol / L Tris-HCl (trishydroxymethylaminomethane hydrochloride), pH 8.0; 20mmol / L EDTA (disodium ethylenediaminetetraacetic acid), pH 8.0; 1.4mol / L NaCl) and 90μl 10% SDS (sodium dodecylbenzenesulfonate) and mix well, put in water bath at 55~60℃ for 1.5h, oscillate and mix once every 10min, centrifuge (12,000rpm) for 15min after 1.5h in water bath, take the supernatant and add Phenol / chloroform / isoamyl alcohol (the volume ratio of phenol, chloroform and isoamyl alcohol is 25:24:1) equal to that of the supernatant, centrifuged (12,000rpm) for 5...
Embodiment 3
[0061] The reaction time that embodiment 3 litchi downy mildew RPA detects
[0062] (1) Genomic DNA extraction of P. litchi Pyrosophthora: The CTAB method in Example 2 was used to extract the genomic DNA of P. litchi Pyrosophthora.
[0063] (2) RPA reaction system: 50 μl reaction system, including 29.5 μl reaction buffer (Rehydration buffer), 2.1 μl each of 10 μM PlRPA-F and biotin-labeled PlRPA-R, 0.6 μl probe 10 μM PlRPALF-P, template DNA to be tested 2.0 μl, 11.2 μl of sterile double distilled water, mix the above components and add to RPA lyophilized enzyme powder, then add 2.5 μl 280mM magnesium acetate (MgAc) and mix by inversion; replace the template DNA with an equal amount of wxya 2 O was used as a negative control.
[0064] (3) RPA reaction: Incubate the reaction system in step (2) at 40°C for 5min, mix the reaction tube again, and continue to incubate at 40°C. The total reaction time is 5min, 10min, 15min, 20min, 25min, 30min, 35min. Among them, 5min, 10min, 15m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com