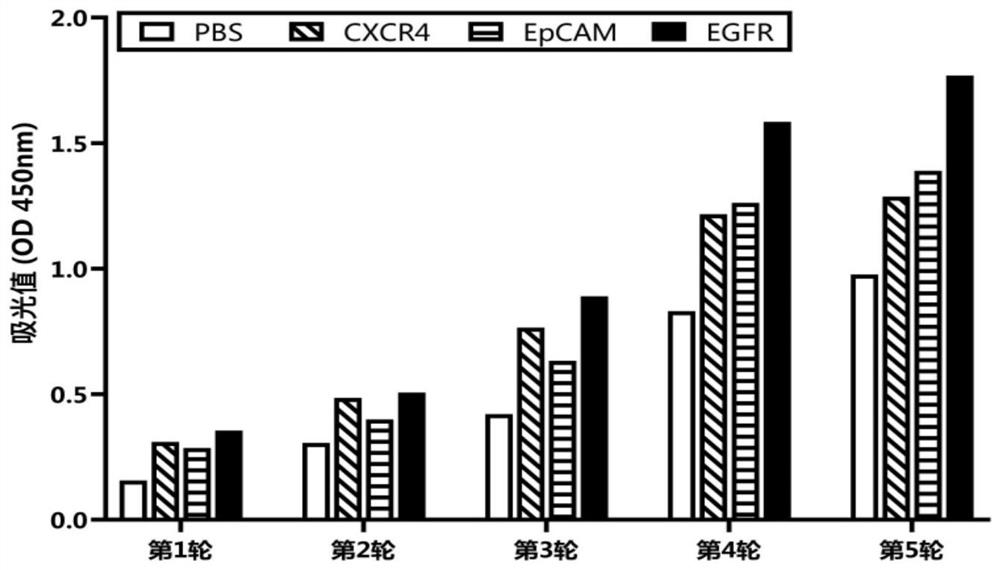
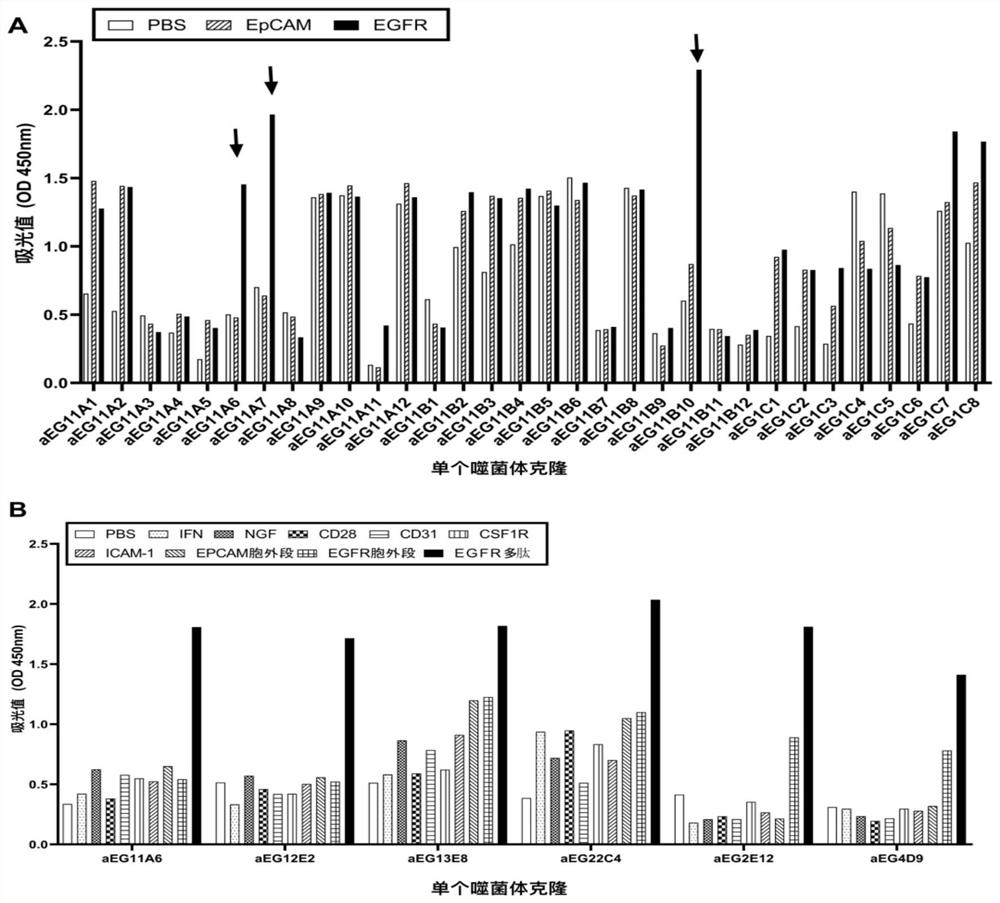
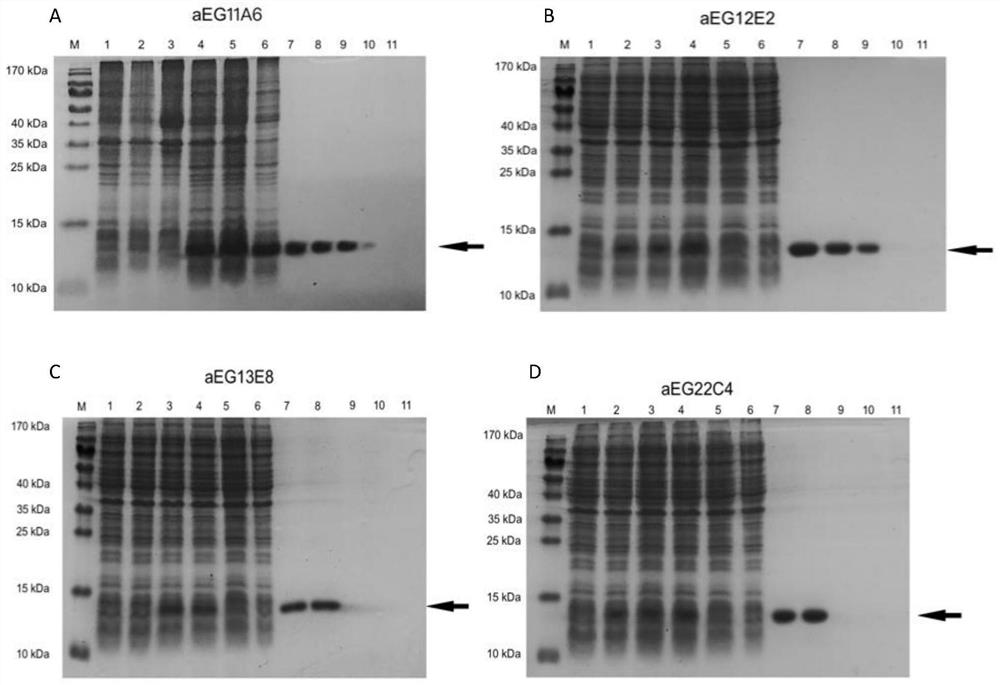
Affinity mature binding protein of EGFR and application
A technology for binding proteins and affinity, applied in the field of affinity mature binding proteins and applications, which can solve the problems of poor permeability, large molecular weight, and inability to effectively remove small lesions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Construction of affinity maturation binding protein library
[0057] (1) Using the wild-type binding protein aEG4D9 (with the nucleotide sequence shown in SEQ ID NO: 12) as a template to construct a mutant library. Mismatch-prone PCR was performed according to the instructions of the GeneMorph II Random Mutagenesis Kit (GeneMorph II, Stratagene). The binding protein DNA is amplified by error-prone PCR and the SfiI restriction site is introduced, and the primers are errorF1 (with the nucleotide sequence shown in SEQ ID NO: 16) and errorR1 (with the nucleotide sequence shown in SEQ ID NO: 17 acid sequences).
[0058] (2) The above PCR products were purified by agarose gel electrophoresis.
[0059] (3) After separating the bands of the correct size, follow Gel Extraction Kit manual operation gel recovery.
[0060] (4) The recovered DNA product is ready for use.
[0061] (5) Add SfiI enzyme to the mismatch-prone PCR product and the pComb3XSS phagemid vector ...
Embodiment 2
[0070] Example 2 preparation of helper phage
[0071] (1) Take out the glycerol-preserved TG1 Escherichia coli clone strain from the -80°C refrigerator, spread it on a TYE plate without antibiotics by the method of four-section line, and culture it upside down overnight at 37°C.
[0072] (2) Pick a TG1 monoclonal strain from the plate and inoculate it into 5 mL of 2×TY (without antibiotics) liquid medium, and culture overnight at 37° C. and 230 rpm.
[0073] (3) Transfer the TG1 bacterial solution to 5 mL of 2×TY antibiotic-free body culture medium at a volume ratio of 1:100, and culture it at 37°C and 230 rpm for 2 hours (bacterial solution OD 600 = around 0.5).
[0074] (4) Take 200 μL TG1 bacterial solution (OD 600 =0.5 or so) into a 1.5mL centrifuge tube, add 10μL KM13 helper phage (1×10 13 pfu / mL), placed in preheated 37°C water bath for 30min.
[0075] (5) Prepare soft agar, let it cool to about 40°C, pour the TG1 bacterial solution treated in step (4), mix, and then...
Embodiment 3
[0080] Example 3 Amplification of affinity maturation binding protein library
[0081] (1) Thaw 5 mL of bacteria containing the binding protein library on ice, transfer to 500 mL of LB liquid medium (containing 0.1% ampicillin by mass volume ratio and 1% glucose by mass volume ratio), and culture in a shaking table at 37°C for 2.5 hours .
[0082] (2) The quantity added is 2×10 12 The pfu helper phage KM13 was incubated at 37°C for 30min.
[0083] (3) Centrifuge at 3200 g for 10 min, and discard the supernatant.
[0084] (4) Resuspend with fresh LB liquid medium (containing 0.1% ampicillin by mass volume ratio and 1% glucose by mass volume ratio). Incubate on a shaker at 25°C for 20 hours.
[0085] (5) Centrifuge at 3200 g for 20 min, and collect the supernatant.
[0086] (6) 20% (w / v) PEG / NaCl solution was added to precipitate the phage in the solution, and placed on ice for 4 hours.
[0087] (7) Centrifuge at 3200 g for 30 min to collect the phage precipitate.
[0088...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Titer | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com