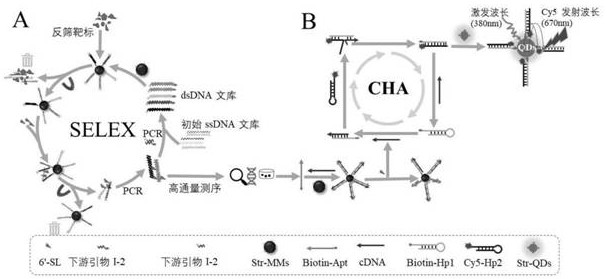
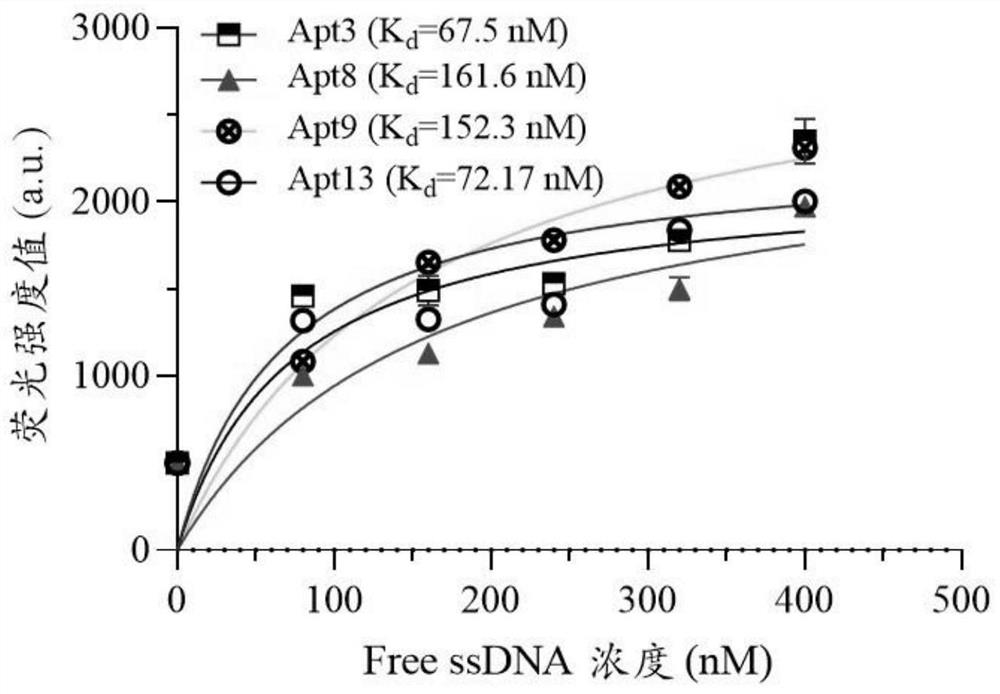
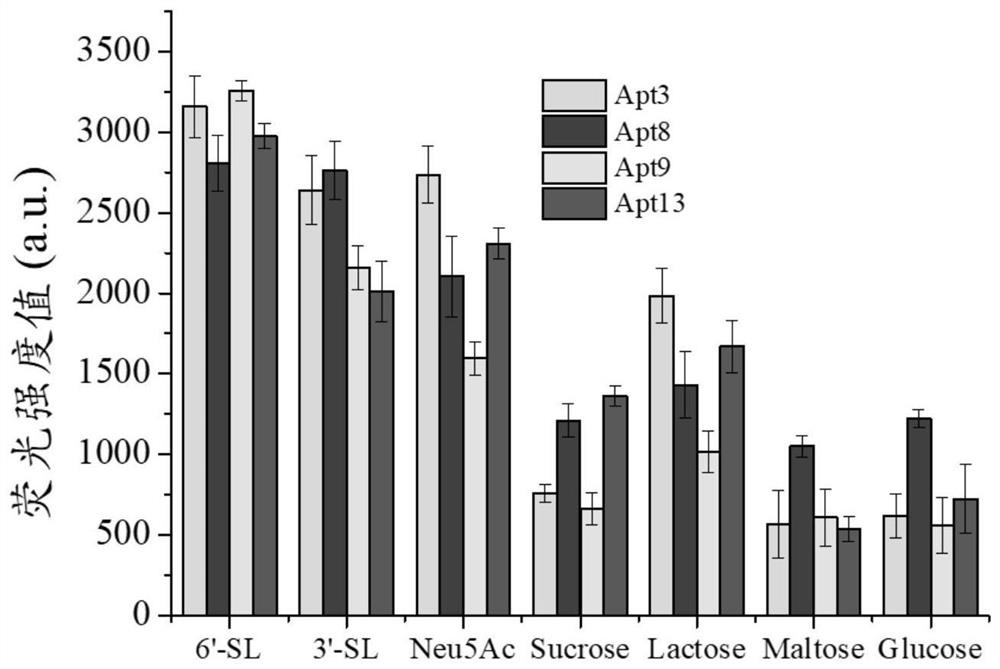
ssDNA aptamer specifically recognizing 6'-sialyllactose and its screening method and application
A sialyllactose, aptamer technology, applied in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problem of poor affinity and specificity between molecularly imprinted polymer and boronate, high cost of antibody preparation, It can not analyze problems such as sugar difference, and achieve the effect of high sensitivity, short operation process and reduced impact.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] (1) Construction of random ssDNA library and its primers:
[0054] a. Construct a random ssDNA library with a length of 79 bases
[0055] 5'-TAGGGAATTCGTCGACGGATCC-N35-CTGCAGGTCGACGCATGCGC CG-3', wherein N represents any one of bases A, T, C, and G.
[0056] b. Synthesis of upstream primers
[0057] Upstream primer 1: 5'-TAGGGAATTCGTCGACGGAT-3';
[0058] Upstream primer 2: 5′-FAM-TAGGGAATTCGTCGACGGAT-3′;
[0059] c. Synthesize downstream primers
[0060] Downstream primer 1: 5'-CGGCGCATGCGTCGACCTG-3';
[0061] Downstream primer 2: 5'-biotin-CGGCGCATGCGTCGACCTG-3'.
[0062] (2) In vitro screening of nucleic acid aptamers:
[0063] S1. Using upstream primers containing fluorescent groups and downstream primers modified with biotin, PCR amplifies the 79nt-length single-stranded DNA library to construct double-stranded DNA for 6′-sialyllactose-specific aptamer screening Library, in which 25 μL of the PCR amplification system is shown in Table 1.
[0064] Table 1 PCR...
Embodiment 2
[0084] S1. Denature the aptamer and the signal probe at 95°C for 5 minutes, then incubate at 37°C for 10 minutes, then add streptavidin-modified magnetic beads and shake slightly for a period of time to form a For the nucleic acid molecular hybridization system of aptamers and signal probes, use a magnet to magnetically separate the solution, discard the supernatant, wash the pellet once with PBS buffer, wash the pellet three times with Tris-HCl buffer, and resuspend In Tris-HCl buffer;
[0085] S2. Add 50 μL of 6′-sialyllactose containing different concentrations to the above Tris-HCl buffer and mix well, and incubate at room temperature for 1 hour; perform magnetic separation with a magnet, and obtain the supernatant, which contains the signal probe. The signal probe sequence is: 5'-CCGTAGTGCGCTCTCGCTCGTGGCT-3';
[0086] S3. Take 200 μL of 1 μmol·L respectively -1 Hairpin probe Hp1 with 200 μL 1 μmol L -1 The hairpin probe Hp2 was denatured at 95°C for 5 minutes, and then...
Embodiment 3
[0095] In order to further verify the accuracy of this method in determining the content of 6′-sialyllactose in actual samples, milk products processed by centrifugal supernatant were selected.
[0096] Take 10mL milk sample and centrifuge at room temperature for 5min (5,000×g), then remove the upper fat layer. Then the milk product was diluted 20 times with Tris-HCl buffer. Then add different concentrations of 6′-sialyllactose and mix well. 5μmol·L -1 The aptamer sequence and the complementary signal probe were denatured at high temperature for 5 minutes, then incubated in ice bath for 10 minutes, and then incubated at 37°C for 1 hour. After that, the supernatant was removed by magnetic separation, and the precipitate was washed once with PBS buffer and Tris-HCl buffer. After washing 3 times, resuspend, mix with 50 μL of milk products containing different concentrations of 6′-sialyllactose, and incubate at room temperature for 30 min with slight shaking. The supernatant wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com