Method for detecting OGT enzyme activity in vitro
An in vitro detection and enzyme activity technology, applied in the field of biochemical analysis and detection, can solve the problems of unfriendly environment, the existence of azide synthesis process, and no activity, etc., and achieve the effect of high detection signal intensity, high conversion efficiency and simple operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
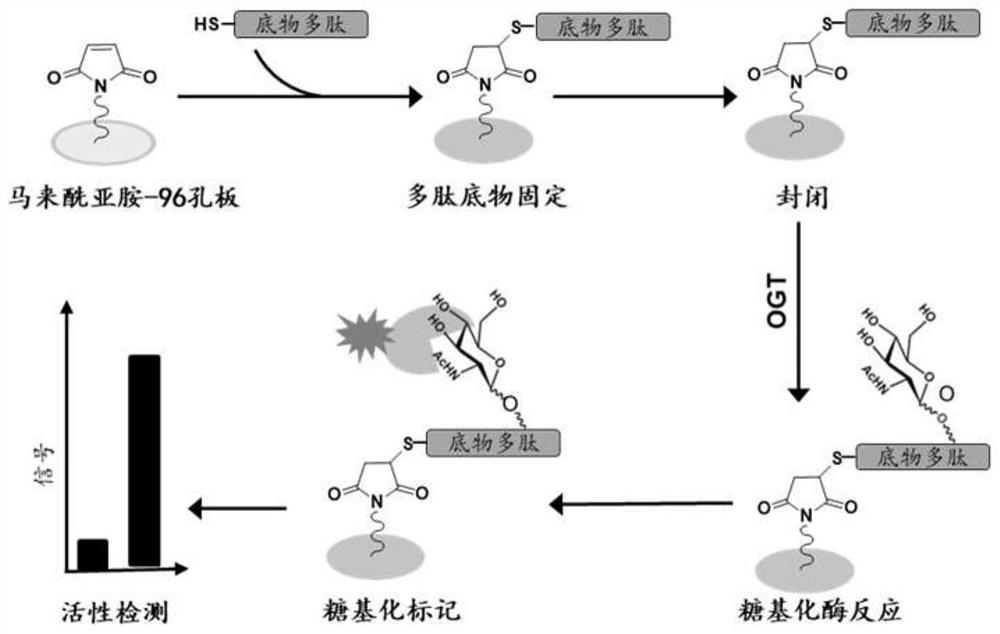
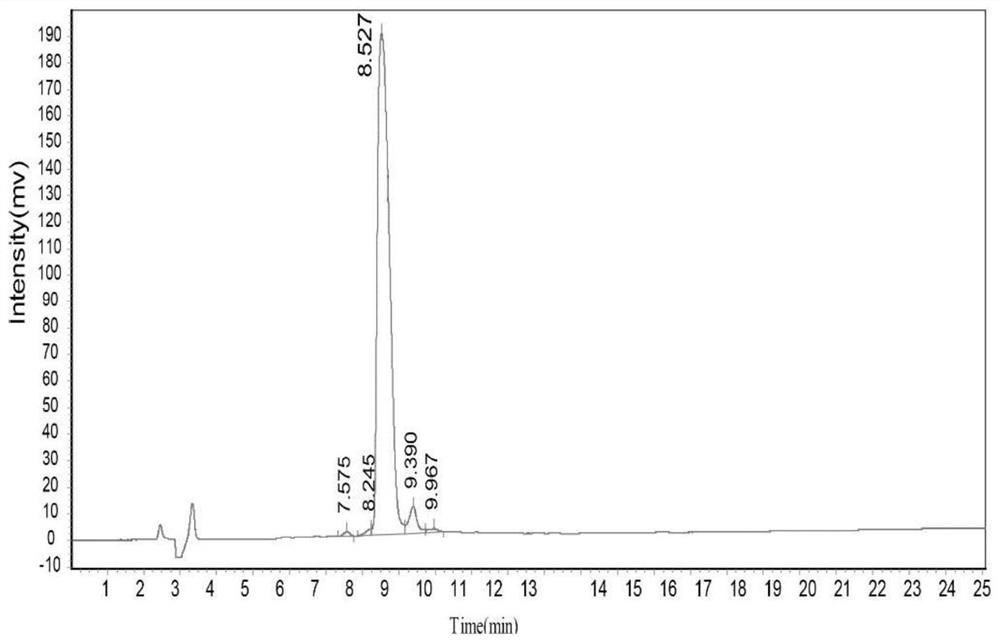
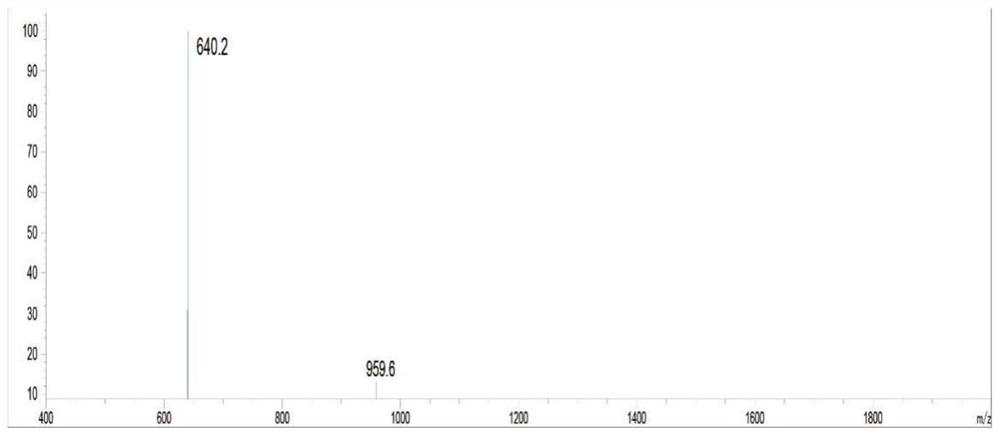
[0068] Example 1 Design and preparation of highly active OGT substrate polypeptide ZO3-S371A
[0069] In this example, the amino acid sequence of the highly active OGT substrate polypeptide ZO3-S371A is derived from the ZO3 protein sequence (NP_001254489.1, SEQ ID No.1) in the NCBI database. Select the amino acid sequence corresponding to positions 357-371 in SEQ ID No.1, change the carboxy-terminal amino acid of the sequence to alanine, and add a glycine and a cysteine to the amino-terminal to obtain the present invention. Highly active OGT substrate polypeptide ZO3-S371A (SEQID No.2) with coupling function. The specific preparation method of ZO3-S371A polypeptide is as follows:
[0070] The preparation of ZO3-S371A polypeptide adopts the principle of polypeptide solid-phase synthesis based on the Fmoc protection strategy, and is synthesized by Symphony polypeptide solid-phase synthesizer. All amino acids constituting the polypeptide of SEQ ID No.2 are standard amino acid...
Embodiment 2
[0075] Example 2 Immobilization and blocking of highly active OGT substrate polypeptide ZO3-S371A
[0076] Dissolve the OGT substrate polypeptide ZO3-S371A in a fixation buffer containing 0.1 mM Na 3 PO 4 , 0.15mM NaCl, 10mM EDTA, pH 7.2, the concentration of the polypeptide substrate after dissolution is 50 ug / ml. The 96-well plate containing activated maleimide groups was purchased from Thermo Fisher Scientific (Cat. No. 15150). Wash the well plate three times with 200ul / well of washing buffer and discard the washing buffer. The wash buffer contains 0.1 mM Na 3 PO 4 , 0.15mM NaCl, 0.05% tween-20, pH 7.2. The polypeptide substrate dissolved in the previous step was incubated with a 96-well plate containing maleimide for 2 hours at a temperature of 25°C. After incubation, the supernatant was discarded, the plate was washed three times with 200ul / well of washing buffer, and the washing buffer was discarded. Configure the blocking solution as 10ug / ml cysteine aqueous sol...
Embodiment 3
[0077] Example 3 Enzymatic reaction of OGT to ZO3-S371A glycosylation
[0078] The buffer system of OGT enzyme reaction is 50mM Tris-HCl, 1mM DTT, 12.5mM MgCl 2 , pH 7.5. The donor substrate for OGT enzyme is UDP-GlcNAc. Add the donor substrate UDP-GlcNAc of OGT to the above-mentioned OGT enzyme reaction system to make its final concentration reach 100um / L, further add purified OGT enzyme 0.5ug or crude enzyme 5ug, and mix well. The above 100ul / well enzyme reaction mixture was added to the 96-well plate immobilized with the OGT substrate polypeptide, the shaking table was rotated at 200rpm, and the temperature was 30°C, and incubated for 3h.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com