Hypothetical protein gene, primer probe group thereof and application of hypothetical protein gene and primer probe group in detection of Meiqi yeast two sharp
A technology of primer probe and hypothetical protein, which is applied in the field of quantitative detection of bicuspid meschis yeast, can solve problems such as not being able to meet production requirements, and achieve the effects of high sensitivity, strong specificity, and high detection accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] The gene fragment (shown as SEQ ID NO. 1) for detecting M. bicides provided by the present invention is derived from the hypothetical protein gene of M. bicides, and the nucleotide sequence of the hypothetical protein gene is as shown in SEQ ID NO. 2 shown. Through the whole genome alignment, it was found that the hypothetical protein gene is a unique gene of M. biloba, so the gene and its gene fragment can be used as a specific detection fragment to distinguish M. michelia from other strains. It is of great significance to improve the specificity and sensitivity of the detection of M. bicides.
[0020] Use the software Primer Primer 5.0 to design specific primers for the hypothetical protein gene (synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.) of M. bicides, marked as:
[0021] HP-F: AAACCCGCAAACTCCACAGA;
[0022] HP-R: TGGATATCACGCTCCATCATTT.
[0023] The strain of M. biloba JMB-1 was cultivated, and total DNA was extracted with a yeast genome extractio...
Embodiment 2
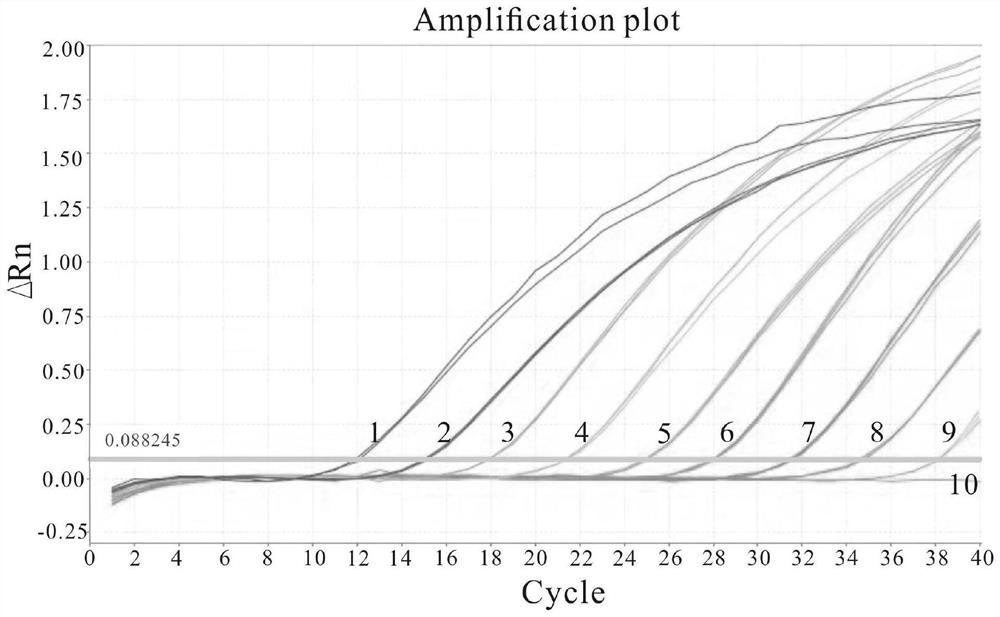
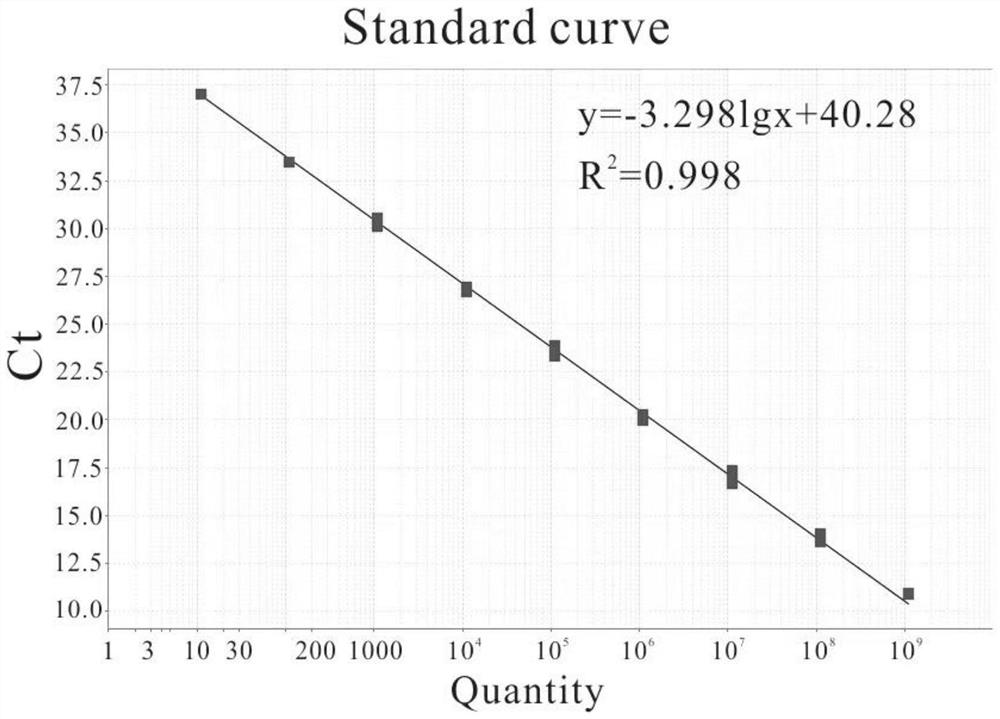
[0025] Using the formula copies / μL=(ng / μL×10 -9 )×(6.02×10 23 ) / (DNA length×660), convert the mass concentration to the copy number concentration according to the molecular weight of the recombinant plasmid standard, and then use sterile water (ddH 2 O) Dilute the recombinant plasmid standard 10-fold serially, and then dilute to 1.13×10 9 Copy / μL~1.13×10 1 copies / μL, a total of 9 concentration gradients.
[0026] Using the serially diluted recombinant plasmid samples as templates, HP-F and HP-R as primers, and HP-TaqMan:FAM-AAATCCACAATACATTGAAGAGCAGCGCA-BHQ1 as probes, real-time quantitative PCR was performed.
[0027] The reaction system of real-time fluorescence quantitative PCR is: 2×Premix Ex Taq (Probe qPCR) 10 μL, upstream detection primer (HP-F) 1 μL, downstream detection primer (HP-R) 1 μL, probe (HP-TaqMan) 0.8 μL, Template 2 μL, sterile water 5.2 μL.
[0028] The amplification procedure was as follows: denaturation at 95°C for 20s; denaturation at 95°C for 1s, a...
Embodiment 3
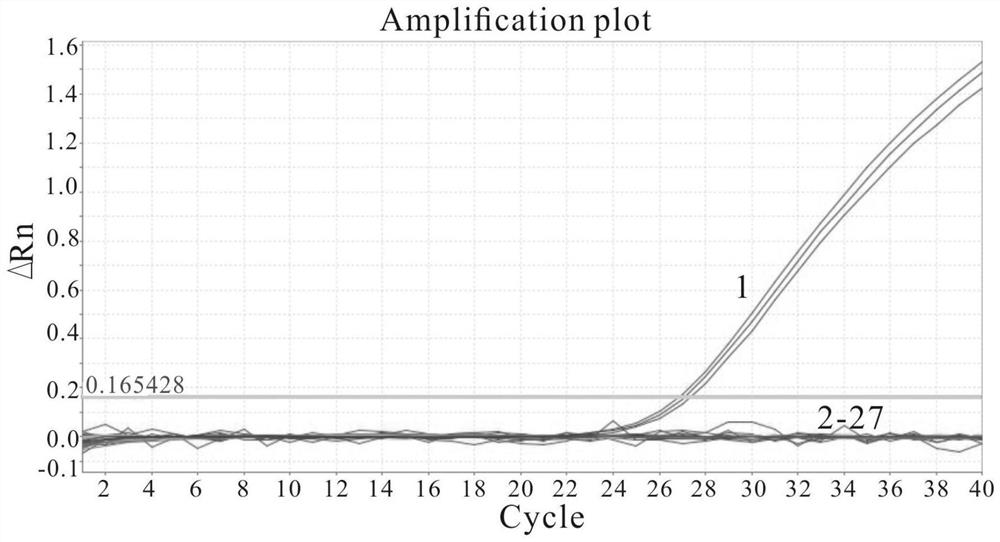
[0031] M. biloba, M. lakoffii, Saccharomyces cerevisiae, Candida tropicalis, Saccharomyces niger, Pichia kudzu, Saccharomyces aureus, Rhododendron, Rhododendron, Hansenula Debaryomyces, Vibrio alginolyticus, Vibrio harveii, Flavobacterium columnar, Streptococcus agalactiae, Streptococcus dysgalactiae, Vibrio eel, Vibrio parahaemolyticus, Photobacterium mermaidii, Aeromonas hydrophila, Aeromonas mildew, Aeromonas viridis, Bacillus cereus, Vibrio vulnificus, Mycobacterium marinum, Nocardia amberjack, Edwardsiella lentus were individually cultured, and DNA extraction kits (purchased from Bao Bioengineering (Dalian) Co., Ltd.) extracted its DNA as a template, and used nuclease-free water (RNase-Free ddH2O) as a negative control, using the primers and probe sets shown in SEQ ID NO. PCR.
[0032] The result is as image 3 As shown, only the samples templated with M. michelia DNA had significant amplification curves. There are no obvious peaks in other bacteria and the negative co...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com