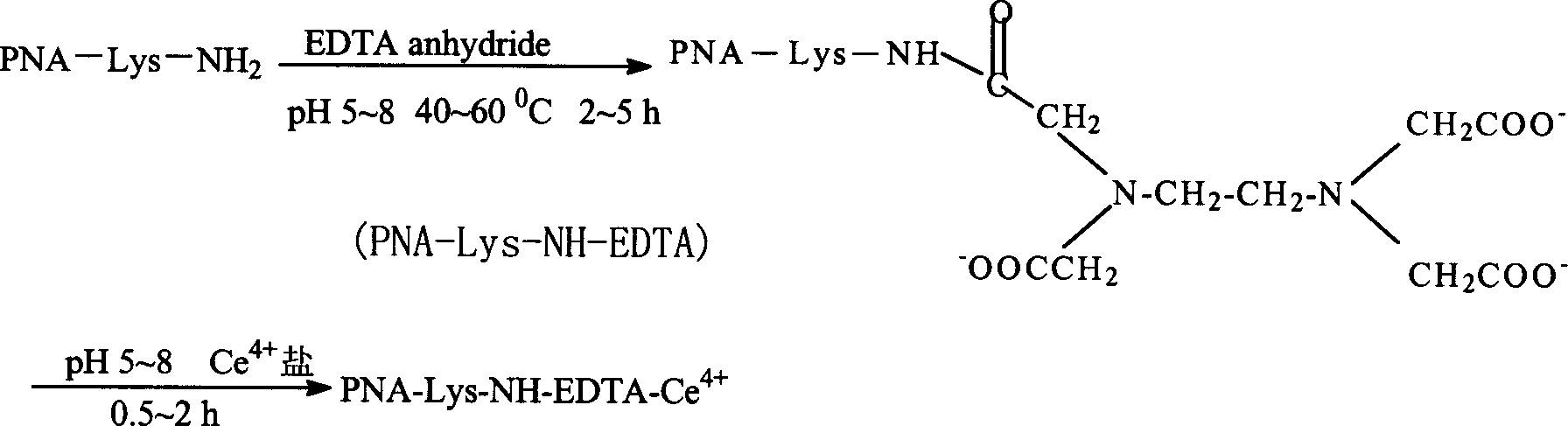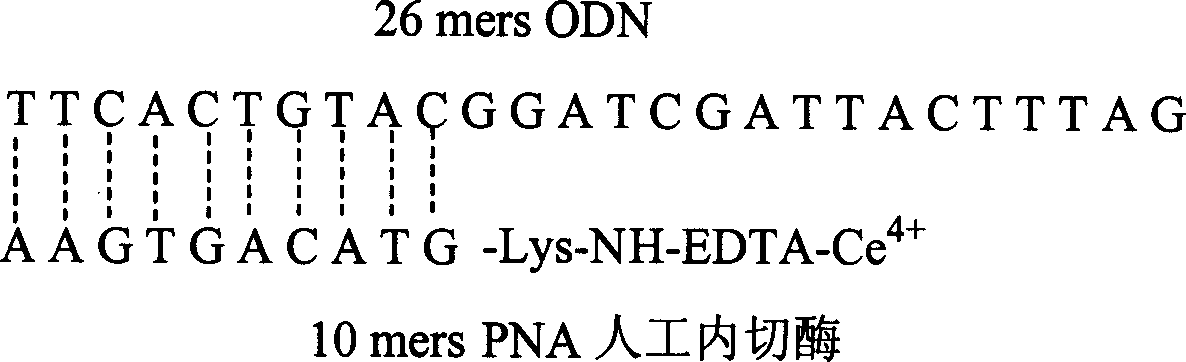Artificial endo enzyme containing peptide nucleic acid recognition element, and its preparing method and use
A technology for recognizing elements and endonucleases, applied in the fields of molecular biology and enzymology, can solve problems such as the lack of DNA substrate specificity or selective cleavage
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0041]Methods for preparing peptide nucleic acids are well known in the art and are described in numerous documents, for example Oliver Seitz, "Solid Phase Synthesis Of Protected Peptide Nucleic Acid" Tetrahedron Letters 40, 4161-4164, 1999 and Birgitte Hyrup et al., "Peptide Nucleic Acids (PNA): Synthesis, Properties and Potential Application", Bioorganic & Medicinal Chemistry, Vol.4, No.1, pp.5-23, 1996.
[0042] In the present invention, the length of the identification element is not particularly limited. Depending on the use of the artificial endonuclease, the length of the recognition element can be at least 4 amino acid structural units, preferably 6-200 structural units, more preferably 8-100 structural units, and most preferably 10- 50 structural units. A particularly preferred recognition element is peptide nucleic acid (abbreviated "PNA"). Due to the development of PCR synthesis technology, DNA synthesis technology and many isolation technologies, those skilled in...
Embodiment 1
[0067] Synthesis of Artificial Endonuclease
[0068] The recognition element is a peptide nucleic acid composed of 10 amino acid structural units, and its sequence is H-GTACAGTGAA-Lys-NH 2 (from N'→C', N' corresponds to the 5' end of the nucleic acid, and C' corresponds to the 3' end of the nucleic acid), purchased from Chengdu Paide Biotechnology Co., Ltd. ) molecule of PNA, forming the PNA-lys-NH 2 .
[0069] With the sequenced PNA, press figure 1 The route shown is for the synthesis of artificial endonucleases. Methods as below:
[0070] (1) Add chelating agent
[0071] To PNA-lys-NH 2 Add Tris-HCl buffer solution with pH=5-8 to dissolve it, then add EDTA anhydride, stir and react at 30°C for 0.5-1h to obtain PNA-lys-NH-EDTA.
[0072] (2) Add cerium ion
[0073] Add Ce ions (Ce(NO 3 ) 4 Form), to obtain PNA-Ce complexes, that is, artificial endonucleases.
Embodiment 2
[0075] positioning cut
[0076] In this example, the ability of the 10-mers PNA-cerium complex artificial nuclease prepared in Example 1 to cleave 26-mers Oligo DNA in a targeted manner was tested.
[0077] If there is a complementary sequence or a homologous sequence part between the PNA single strand and the DNA single strand, it can form a hybrid double strand under annealing conditions through molecular hybridization. The 10 bases in the 10mers PNA-lys-NH-EDTA structure are complementary to the 10 bases in the 26-mers ODN (sequence 5'-TTCACTGTACGGATCGATTACTTTAG-3'), so under appropriate conditions, the bases can be complementary Pairing principle, hybridization forms a double helix structure, such as figure 2 shown.
[0078] The 26-mer oligonucleotide substrate and the artificial endonuclease of Example 1 were reacted in Tris-HCl buffer solution with pH=6-8 at a temperature of 30°C-40°C for 24-48 hours. Then the reaction product was taken for electrophoresis.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com