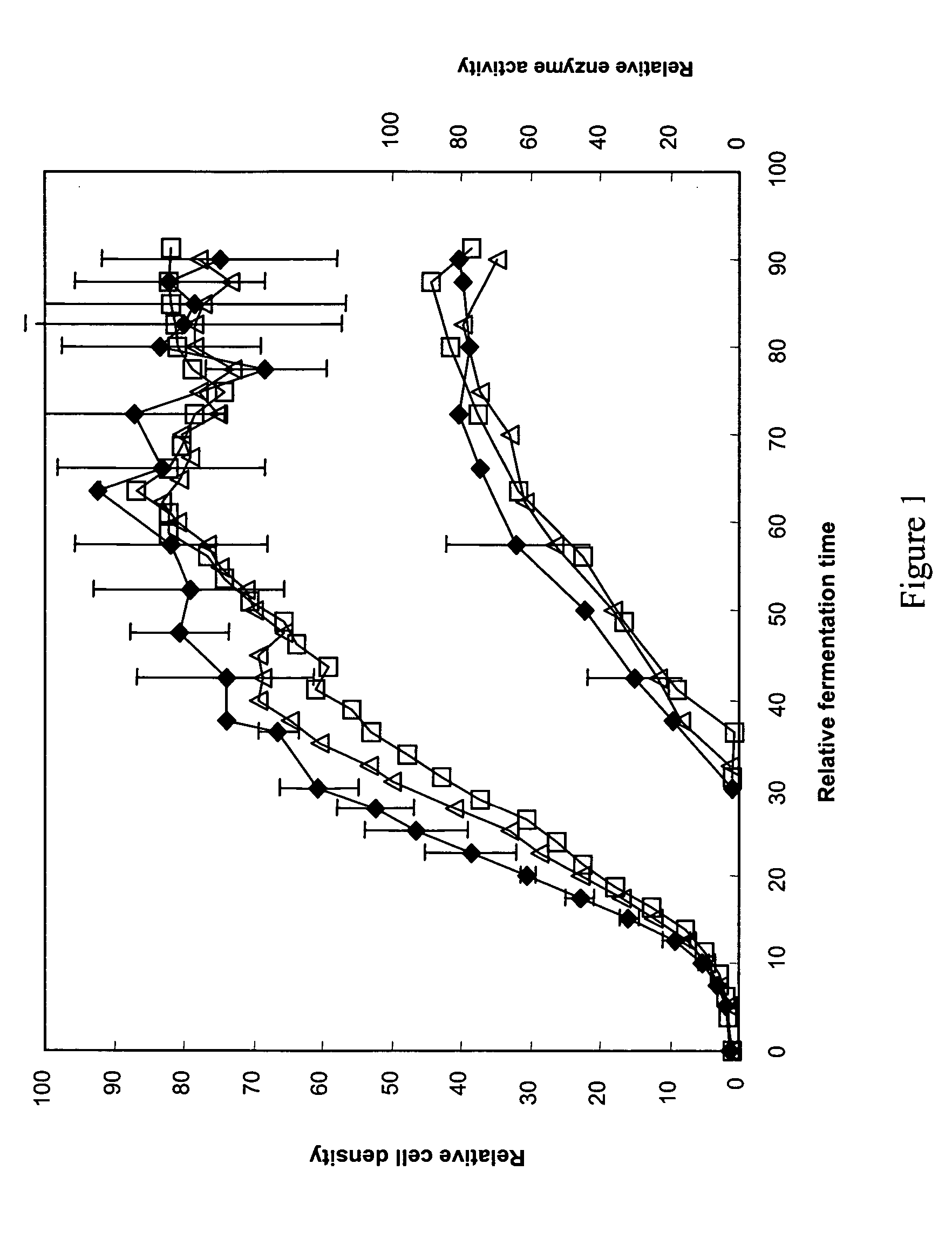
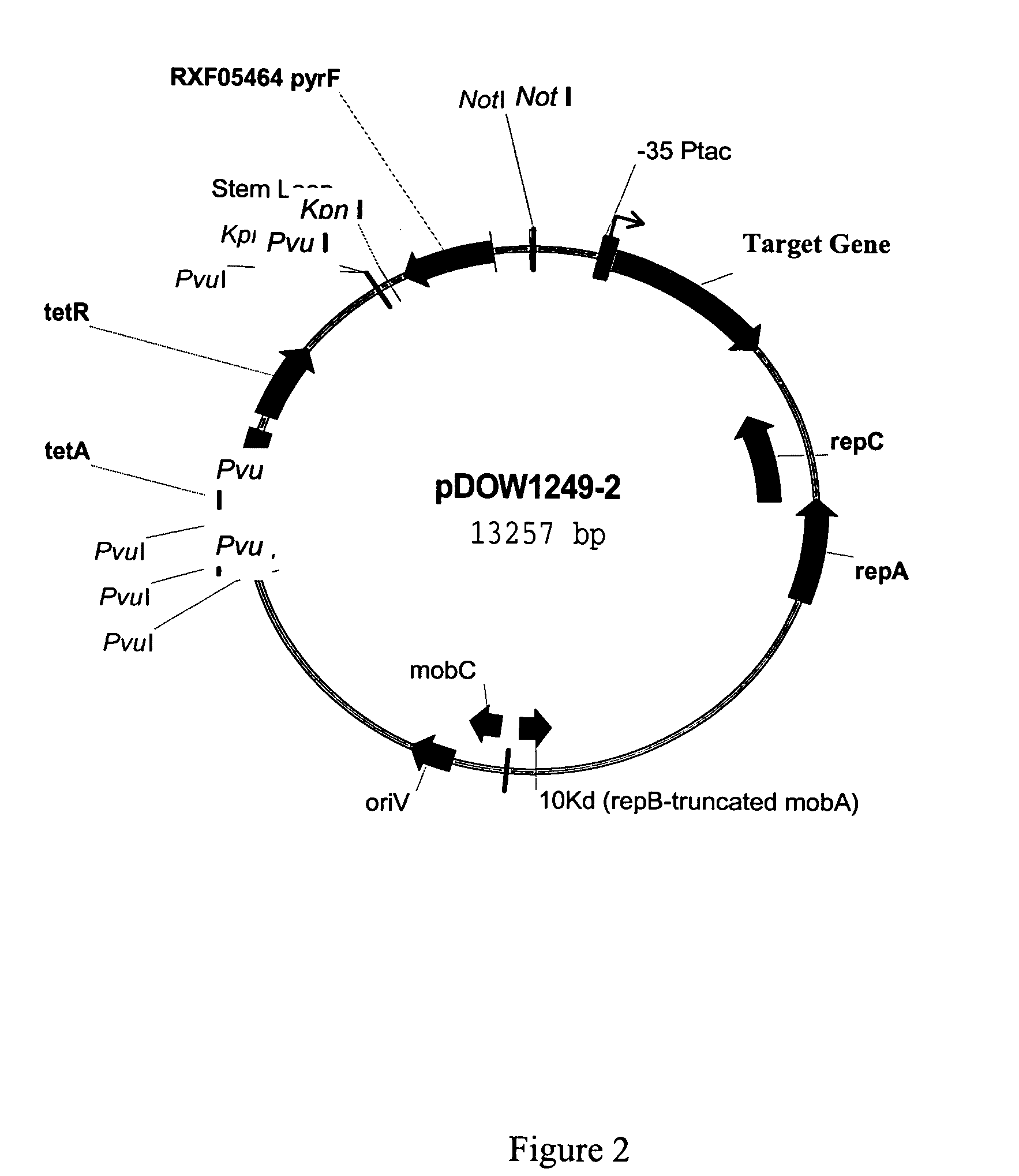
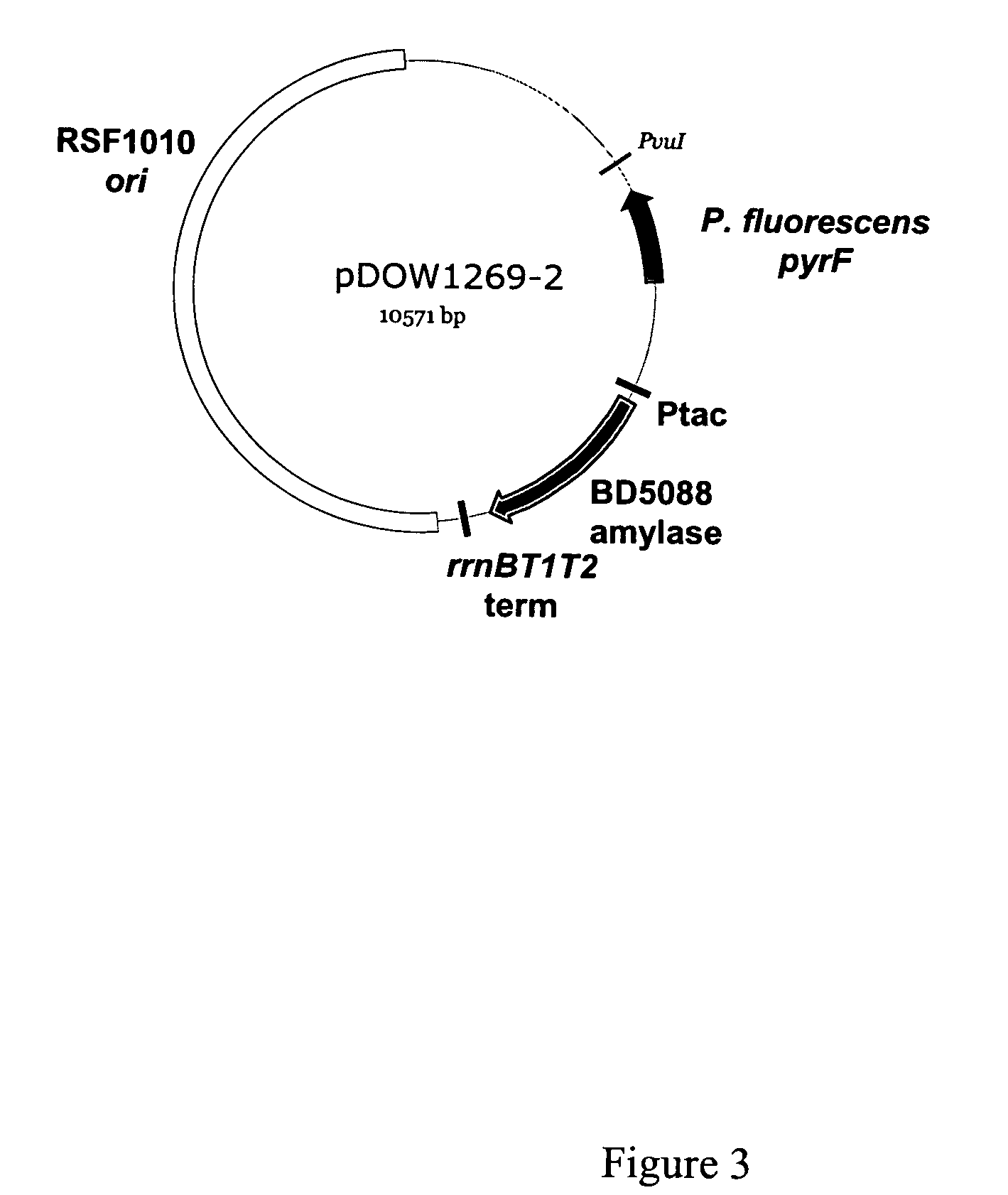
Protein expression systems
a protein and expression system technology, applied in the field of protein expression systems, can solve the problems of reducing the fermentation efficiency of the production of the desired recombinant, unable to survive host cells lacking the selection marker, such as revertants, and unable to achieve the production of superior host cells. , to achieve the effect of improving the production of bacteria's protein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of a pyrF Selection Marker System in a P. fluorescens Host Cell Expression System
[0279] Reagents were acquired from Sigma-Aldrich (St. Louis Mo.) unless otherwise noted. LB is 10 g / L tryptone, 5 g / L yeast extract and 5 g / L NaCI in a gelatin capsule (BIO 101). When required, uracil (from BIO101, Carlsbad Calif.) or L-proline was added to a final concentration of 250 ug / mL, and tetracycline was added to 15 ug / mL. LB / 5-FOA plates contain LB with 250 mM uracil and 0.5 mg / mL 5-fluoroorotic acid (5-FOA). M9 media consists of 6 g / L Na2HPO4, 3 g / L KH2PO4, 1 g / L NH4Cl, 0.5 g / L NaCl, 10 mM MgSO4, 1× HoLe Trace Element Solution, pH7. Glucose was added to a final concentration of 1%. The 1000× HoLe Trace Element Solution is 2.85 g / L H3BO3, 1.8 g / L MnCl2 . 4H2O, 1.77 g / L sodium tartrate, 1.36 g / L FeSO4. 7H2O, 0.04 g / L CoCl2. 6H2O, 0.027 g / L CuCl2. 2H2O, 0.025 g / L Na2MoO4. 2H2O, 0.02 g / L ZnCl2.
Oligonucleotides Used Herein
[0280]
MB214pyrF1 (NotI site in bold)5′-GCGGCCGCTTTGGCGCTTCGT...
example 2
Construction of a pyrF—proC Dual Auxotrophic Selection Marker System in a P. fluorescens Host Cell Expression System
[0292] Oligonucleotides Used Herein
proC15′-ATATGAGCTCCGACCTTGAGTCGGCCATTG-(SEQ ID NO: 22)3′proC25′-ATATGAGCTCGGATCCAGTACGATCAGCAGG(SEQ ID NO: 23)TACAG-3′proC35′-AGCAACACGCGTATTGCCTT-3′(SEQ ID NO: 24)proC55′-GCCCTTGAGTTGGCACTTCATCG-3′(SEQ ID NO: 25)proC65′-GATAAACGCGAAGATCGGCGAGATA-3′(SEQ ID NO: 26)proC75′-CCGAGCATGTTTGATTAGACAGGTCCTTATT(SEQ ID NO: 27)TCGA-3′proC85′-TGCAACGTGACGCAAGCAGCATCCA-3′(SEQ ID NO: 28)proC95′-GGAACGATCAGCACAAGCCATGCTA-3′(SEQ ID NO: 29)genF25′-ATATGAGCTCTGCCGTGATCGAAATCCAGA-(SEQ ID NO: 30)3′genR25′-ATATGGATCCCGGCGTTGTGACAATTTACC-(SEQ ID NO: 31)3′XbaNotDraU2 linker5′-TCTAGAGCGGCCGCGTT-3′(SEQ ID NO: 32)XbaNotDraL linker5′-GCGGCCGCTCTAGAAAC-3′(SEQ ID NO: 33)
Cloning of proC from P. fluorescens and Formation of a pCN Expression Plasmid Containing proC
Replacing Antibiotic Resistant Gene in pCN51lacI with proC
[0293] The proC ORF and about 100 bp of...
example 3
Chromosomal Integration of lacI. lacIQ and lacIQ1 in P. fluorescens
[0316] Three P. fluorescens strains have been constructed, each with one of three different Escherichia coli lacI alleles, lacI (SEQ ID NO:9), lacIQ (SEQ ID NO: 11), and lacIQ1 (SEQ ID NO:12), integrated into the chromosome. The three strains exhibit differing amounts of LacI repressor accumulation. Each strain carries a single copy of its lacI gene at the levansucrase locus (SEQ ID NO:13) of P. fluorescens DC36, which is an MB101 derivative (see TD Landry et al., “Safety evaluation of an α-amylase enzyme preparation derived from the archaeal order Thermococcales as expressed in Pseudomonas fluorescens biovar I,”Regulatory Toxicology and Pharmacology 37(1): 149-168(2003)) formed by deleting the pyrF gene thereof, as described above.
[0317] No vector or other foreign DNA sequences remain in the strains. The strains are antibiotic-resistance-gene free and also contain a pyrF deletion, permitting maintenance, during gr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com