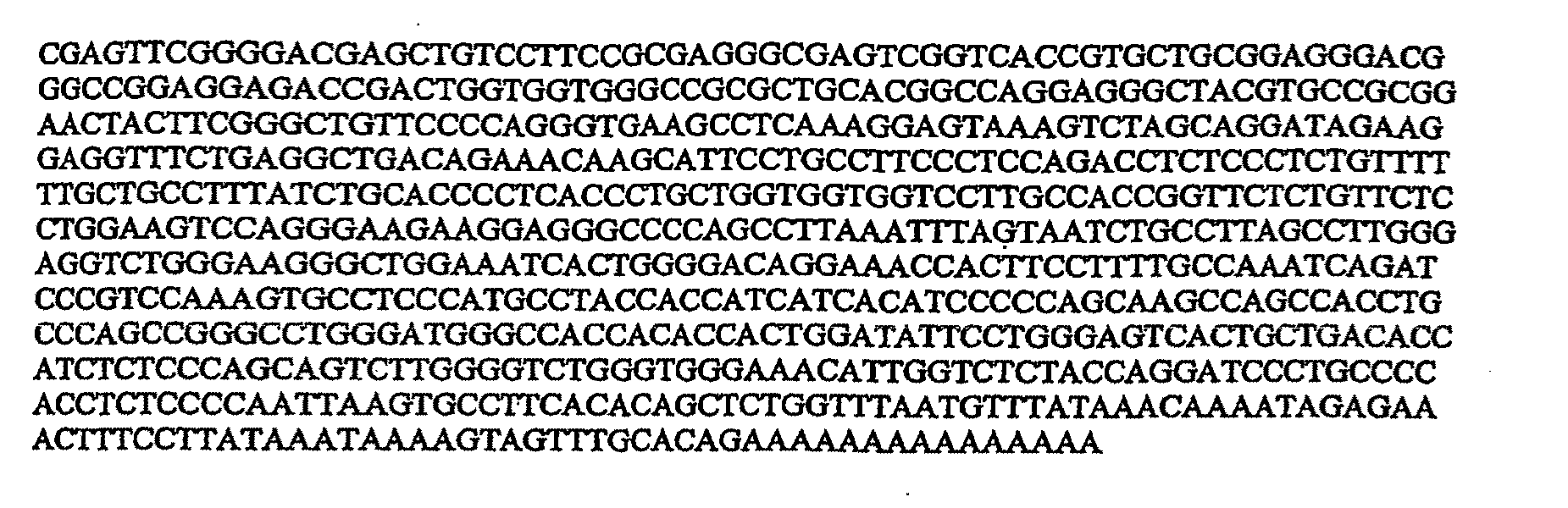[0027]Adaptations which facilitate the expression of vector encoded genes include the provision of transcription termination /
polyadenylation sequences. This also includes the provision of internal
ribosome entry sites (IRES) which function to maximise expression of vector encoded genes arranged in bicistronic or multi-cistronic expression cassettes.
[0043]A modified
antibody, or variant
antibody and reference
antibody, may differ in
amino acid sequence by one or more substitutions, additions, deletions, truncations which may be present in any combination. Among preferred variants are those that vary from a reference polypeptide by conservative
amino acid substitutions. Such substitutions are those that substitute a given
amino acid by another amino acid of like characteristics. The following non-limiting
list of amino acids are considered conservative replacements (similar): a)
alanine,
serine, and
threonine; b)
glutamic acid and asparatic acid; c)
asparagine and
glutamine d)
arginine and
lysine; e)
isoleucine,
leucine,
methionine and
valine and f)
phenylalanine,
tyrosine and
tryptophan. Most highly preferred are variants which show enhanced
biological activity.
[0053]The therapeutics of the invention can be administered by any conventional
route, including injection or by gradual infusion over time. The administration may, for example, be oral, intravenous, intraperitoneal, intramuscular, intracavity, subcutaneous, or transdermal. When antibodies are used therapeutically, a preferred
route of administration is by pulmonary
aerosol. Techniques for preparing
aerosol delivery systems containing antibodies are well known to those of skill in the art. Generally, such systems should utilize components which will not significantly impair the biological properties of the antibodies, such as the
paratope binding capacity (see, for example, Sciarra and Cutie, “Aerosols,” in Remington's
Pharmaceutical Sciences, 18th edition, 1990, pp 1694-1712; incorporated by reference). Those of skill in the art can readily determine the various parameters and conditions for producing antibody aerosols without resort to undue experimentation. When using antisense preparations of the invention, slow intravenous administration is preferred.
[0068]As used herein, the term “antisense molecule” or “antisense” describes an
oligonucleotide that is an oligoribonucleotide, oligodeoxyribonucleotide, modified oligoribonucleotide, or modified oligodeoxyribonucleotide which hybridises under physiological conditions to
DNA comprising a particular
gene or to an mRNA transcript of that
gene and, thereby, inhibits the transcription of that
gene and / or the translation of that mRNA. The antisense molecules are designed so as to interfere with transcription or translation of a
target gene upon hybridisation with the
target gene or transcript. Those skilled in the art will recognise that the exact length of the antisense
oligonucleotide and its degree of complementarity with its target will depend upon the specific target selected, including the sequence of the target and the particular bases which comprise that sequence. It is preferred that the antisense
oligonucleotide be constructed and arranged so as to bind selectively with the target under physiological conditions, i.e., to hybridise substantially more to the target sequence than to any other sequence in the target
cell under physiological conditions. Based upon the iASPP6C
nucleic acid sequences provided herein, or upon allelic or homologous genomic and / or cDNA sequences, one of skill in the art can easily choose and synthesise any of a number of appropriate antisense molecules for use in accordance with the present invention. For example, a “gene walk” comprising a series of oligonucleotides of 15-30 nucleotides spanning the length of iASPP6C nucleic acid can be prepared, followed by testing for inhibition of the corresponding iASPP6C expression. Optionally, gaps of 5-10 nucleotides can be left between the oligonucleotides to reduce the number of oligonucleotides synthesised and tested.
[0099]It will be apparent to one skilled in the art that modification to the amino acid sequence of peptides agents could enhance the binding and / or stability of the
peptide with respect to its target sequence. In addition, modification of the
peptide may also increase the
in vivo stability of the
peptide thereby reducing the effective amount of peptide necessary to inhibit p53 binding of iASPP. This would advantageously reduce undesirable side effects which may result
in vivo. Modifications include, by example and not by way of limitation,
acetylation and amidation. Alternatively or preferably, said modification includes the use of modified amino acids in the production of recombinant or synthetic forms of peptides. It will be apparent to one skilled in the art that modified amino acids include, by way of example and not by way of limitation, 4-
hydroxyproline, 5-hydroxylysine, N6-acetyllysine, N6-
methyllysine, N6,N6-dimethyllysine, N6,N6,N6-trimethyllysine, cyclohexyalanine, D-amino acids,
ornithine. Other modifications include amino acids with a C2, C3 or C4
alkyl R group optionally substituted by 1, 2 or 3 substituents selected from halo (eg F, Br, I), hydroxy or C1-C4 alkoxy. Modifications also include, by example and not by way of limitation,
acetylation and amidation.
 Login to View More
Login to View More