General multiplex polymerase chain reaction realization method based on microarray chip
A microarray chip and chain reaction technology, applied in the field of biomedicine, can solve the problems that the instrument cannot be applied on a large scale, the maximum multiplicity is low, and the operation is complicated.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Synchronous amplification of 9 exons of Duchenne muscular dystrophy (DMD) gene
[0034] The sample is the blood of a healthy person, and the genomic DNA is extracted using the rapid DNA extraction and amplification kit of Tiangen Biochemical Technology Co., Ltd. This embodiment includes the following steps:
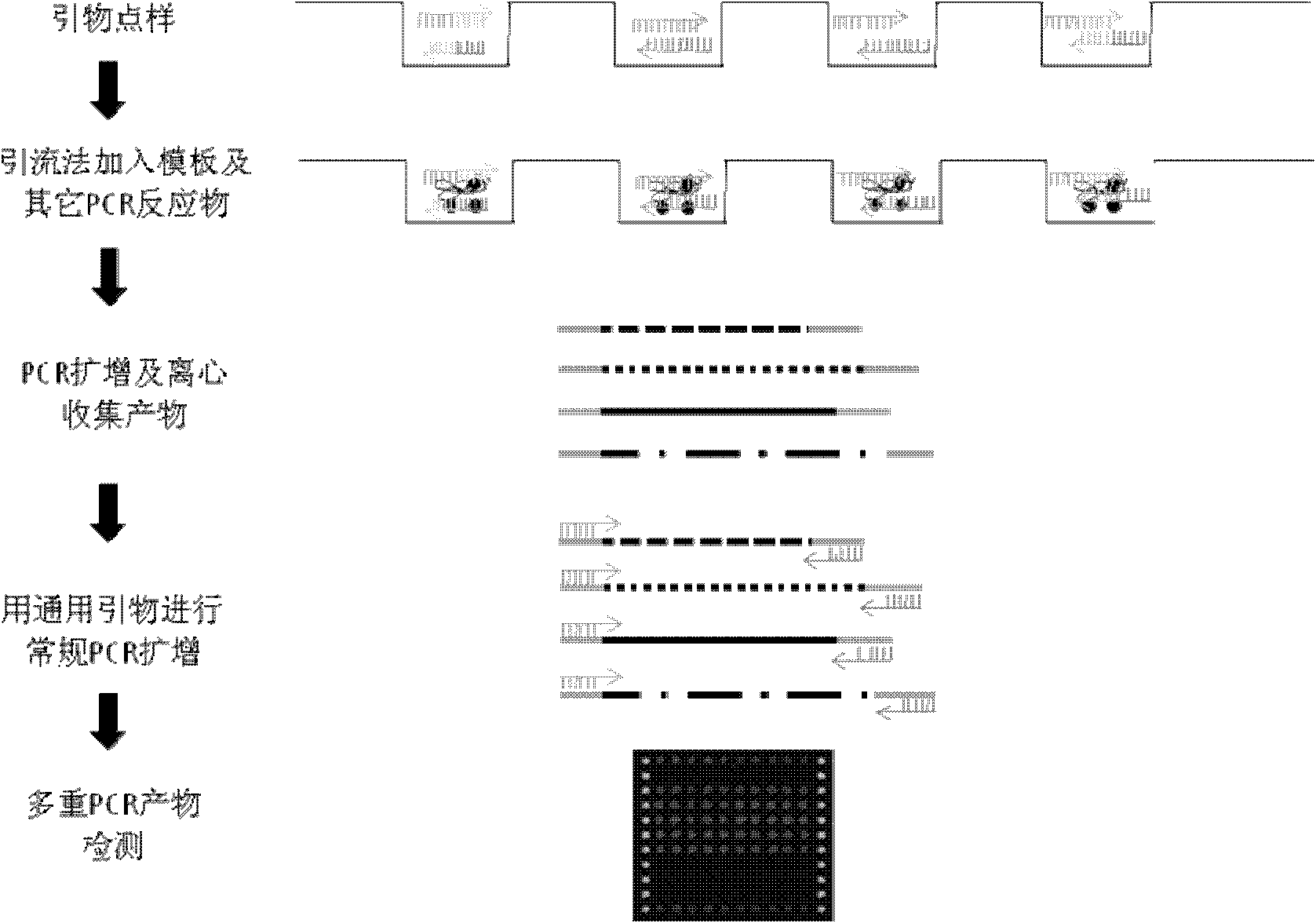
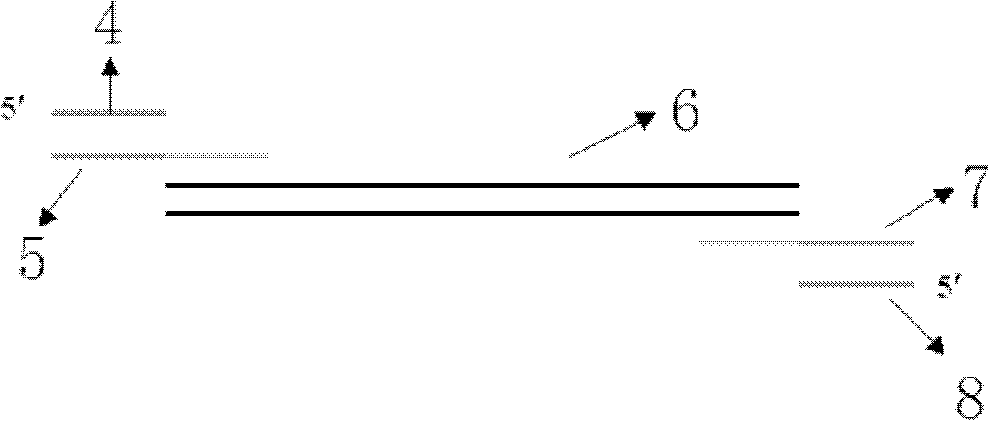
[0035] The first step is to use the microarray chip to spot multiple pairs of primers into several hydrophilic microwells, and then add the PCR components to the hydrophilic microwells by pulling or draining;
[0036] The microarray chip spotting sample comes from Boao Biotechnology Co., Ltd., and the model is SmartArrayer TM 48.
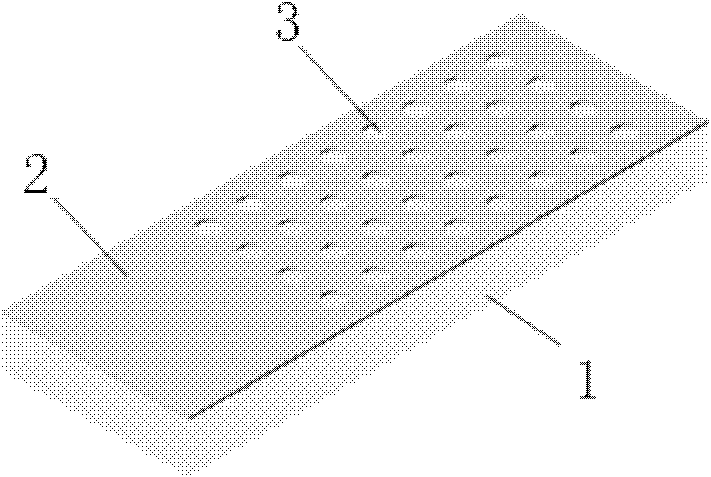
[0037] Such as figure 2 As shown, the microarray chip includes: a substrate, a hydrophobic surface and micropores, wherein: the hydrophobic surface is located on the top of the substrate, and several micropores are arranged through the hydrophobic surface in an array.
[0038] The microarray chip is prepared in the follow...
Embodiment 2
[0059] Example 2: Multiplex amplification and chip detection of 11 kinds of pneumonia-causing cloning plasmids
[0060] 1. Sample:
[0061] The cloning plasmids corresponding to 11 kinds of pneumonia pathogenic bacteria (see the table below) were used as templates. The vector is pGM-T from Tiangen Biochemical Technology Co., Ltd. Plasmids were extracted using a plasmid extraction kit (Tiangen Biochemical).
[0062] Numbering
initials
1
Acinetobacter baumannii
Aba
adeS
2
Streptococcus pneumoniae
Spn
lyA
3
Staphylococcus aureus
Sau
femA
4
Pseudomonas aeruginosa
Pae
opI
5
Sma
wxya
6
influenza bacilli
Hin
ompP6
7
Legionella pneumophila
Lpn
mip
8
Chlamydia pneumoniae
Cpn
PstI
9
Mycoplasma pneumoniae
FH
P1
10
Klebsiella...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com