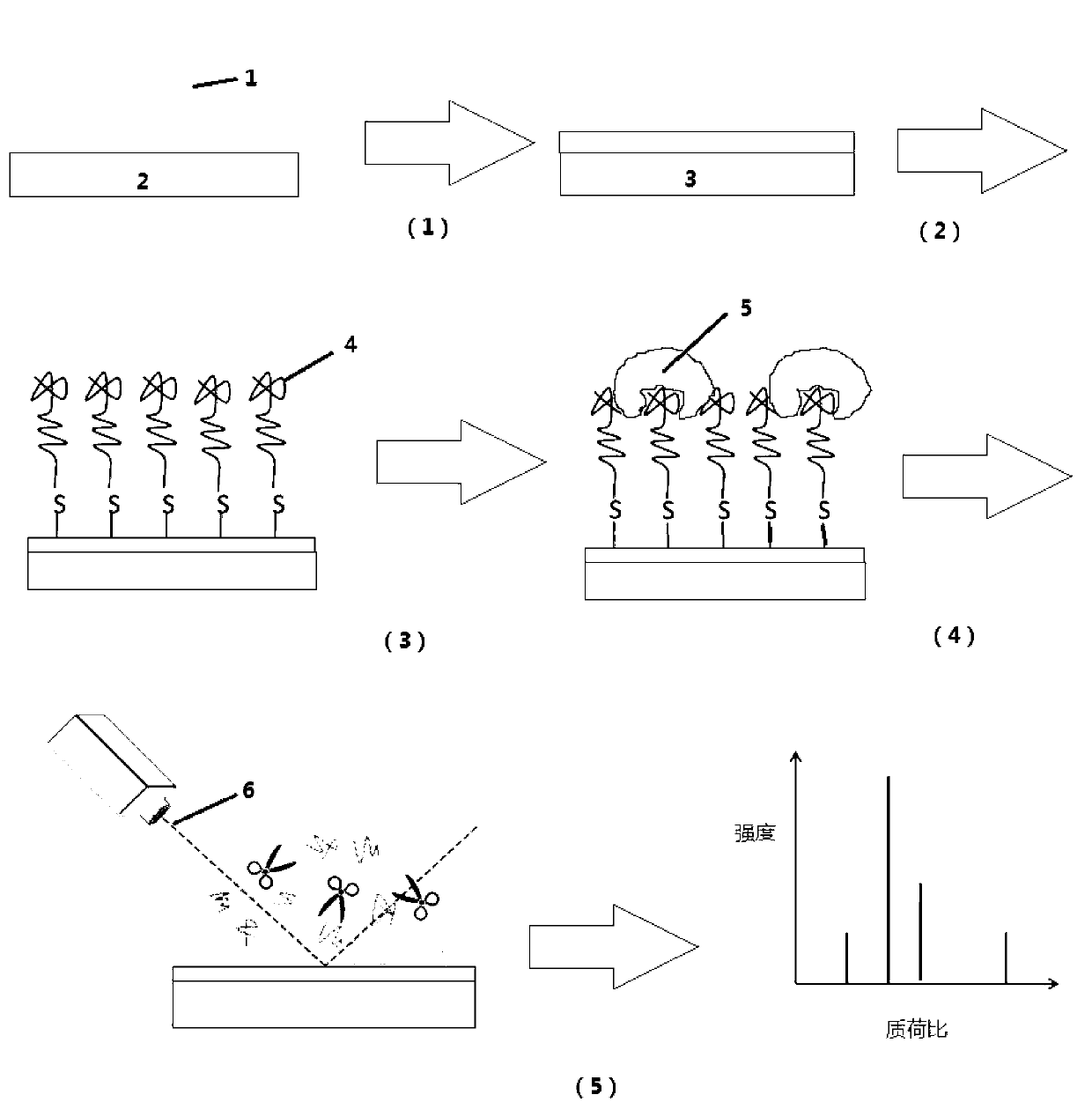
Detection method for protein with nucleic acid aptamer target gathering and laser enzymolysis
A nucleic acid aptamer and protein technology, applied in the field of chemical biology, can solve the problems of difficult detection and low concentration, and achieve the effect of simple method, accurate qualitative and quantitative, and large specific surface area
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1:
[0041] The preparation of the target plate with surface modified gold film, the preparation process is as follows figure 1 Shown:
[0042] (1) Using the classic Frens method to synthesize gold colloid under the protection of sodium citrate, the specific steps are:
[0043] (a) Add 1mmol / L chloroauric acid (HAuCl 4 ) solution and boil;
[0044] (b) Add 38.8mmol / L trisodium citrate solution, and reflux for 15min;
[0045] (c) cooling. The obtained red solution was filtered through a 0.22 μm filter membrane to obtain gold nanocolloids with a size of 13 nm.
[0046] (2) Take 10 mL of gold gel, centrifuge at 20,000 rcf for 20 min, and discard the supernatant. Adjust the gold colloid concentration to 92 nmol / L according to the absorbance at 260 nm;
[0047] (3) Spot 2 μL 92 nmol / L gold glue on a clean target plate and let it dry naturally overnight;
[0048] (4) Adjust the oven to 200 °C, put in the target plate, and calcinate for 2 h;
[0049] (5) Cool...
Embodiment 2
[0051] Example 2: Using lysozyme as a model protein, nucleic acid aptamers were used for on-target enrichment, laser enzymatic hydrolysis and MALDI-TOF MS qualitative and quantitative analysis:
[0052] (1) Immobilization of the thiol-modified lysozyme aptamer: The thiol-modified lysozyme aptamer was treated with 2.5 mM tris(2-carboxyethyl)phosphine (TCEP) for 1 h, and 2 μL was spotted on the target plate. Put the target plate in a wet box overnight in the dark;
[0053] (2) Configuration of standard protein solution: Dilute lysozyme protein to 5000 ng / mL, 1000 ng / mL, 100 ng / mL, 14 ng / mL;
[0054] (3) Enrichment: pipette 2 μL of lysozyme protein solution onto the MALDI target plate, and react in a wet box at room temperature for 1 hour. Rinse with 0.1% Tween 20 and deionized water for 1 min. dry;
[0055] (4) Laser enzymolysis: Prepare 25 ng / μL trypsin solution with 25 mmol / L ammonium bicarbonate buffer. Take 2 μL and spot on the target plate enriched with lysozyme. Irradi...
Embodiment 3
[0058] Example 3: Adding lysozyme to human urine, using nucleic acid aptamers for on-target enrichment, laser enzymolysis and MALDI-TOF MS qualitative and quantitative analysis:
[0059] (1) Preparation of urine samples: Collect morning urine from healthy people, centrifuge to remove precipitate, filter with 0.22 μm filter membrane, and store at -20°C. Add lysozyme directly to the urine sample to concentrations of 5000 ng / mL, 500 ng / mL, and 100 ng / mL;
[0060] (2) According to steps (3), (4) and (5) of Example 2, enrichment, laser enzymatic hydrolysis and mass spectrometry were performed on the sample. Qualitative and quantitative analysis of lysozyme;
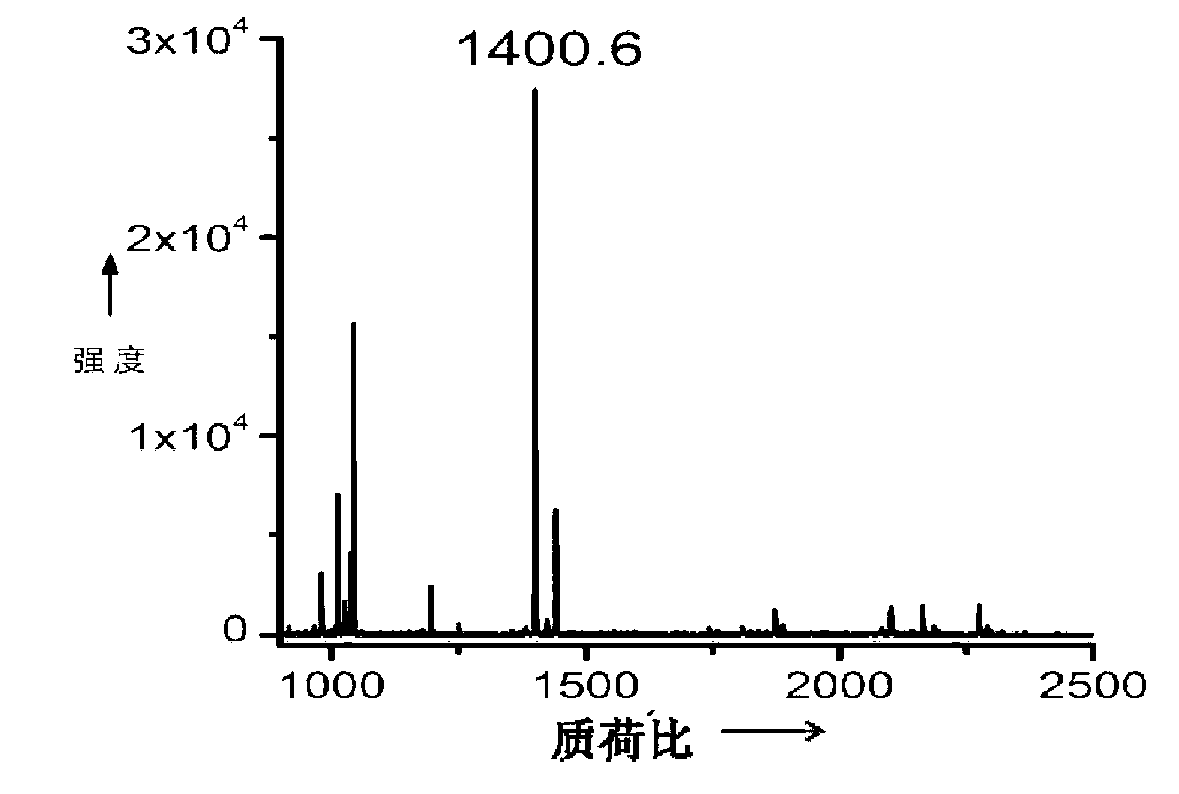
[0061] (3) Detection of endogenous lysozyme in urine: No protein was added to the urine sample, and the blank urine sample was directly enriched as described in Steps (3), (4) and (5) of Example 2, and laser Enzymatic digestion and mass spectrometry analysis. MS / MS analysis was performed on the peptide m / z=1400.6, and compa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com