Ocean cold-adapted esterase as well as coding gene E40 and application thereof
A technology of cold-adapted esterase and cold-adapted ester, which is applied in marine cold-adapted esterase and its coding gene E40 and its application in the field of biology to achieve the effect of ensuring safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] A marine cold-adapted esterase gene E40, the nucleotide sequence of which is shown in SEQ ID NO.1. The above gene encodes a marine cold-adapted esterase E40, the amino acid sequence of which is shown in SEQ ID NO.2.
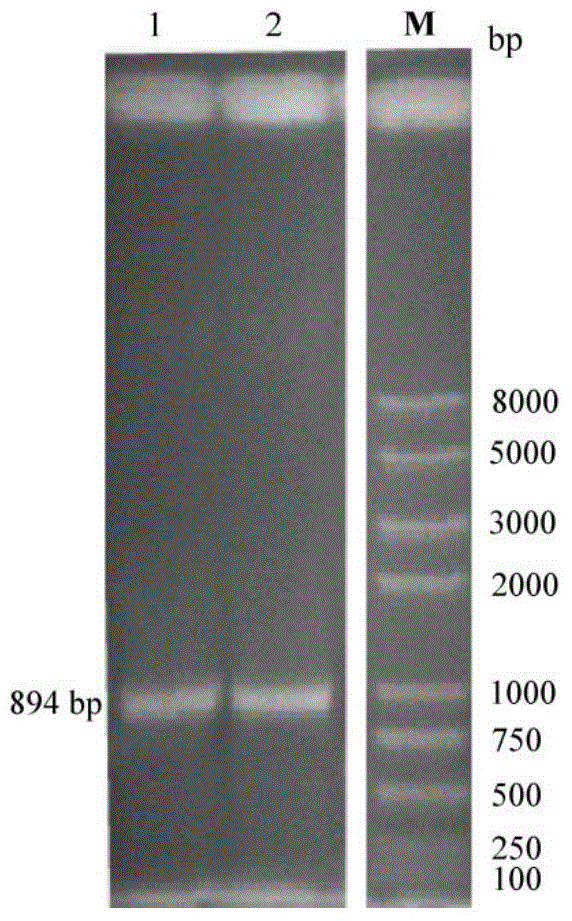
[0032] The gene E40 is 894bp in total, which contains an open reading frame of 894bp, which encodes cryogenic esterase E40, the start codon is located at 1bp, and the stop codon is located at 892bp, encoding a total of 297 amino acids.
Embodiment 2
[0033] Example 2: Determination of the gene sequence encoding cold-adapted esterase E40
[0034] The source of the strain: Escherichia coli EPI300 clone E40-6B in the E505 metagenomic library of the South China Sea seabed sediment sample.
[0035] Specific steps are as follows:
[0036] 1.1 For the extraction of the large fragment plasmid fosmid in E. coli EPI300 clone E40-6B, refer to the instructions of the BAC / PAC DNA extraction kit from OMEGA Company
[0037] (1) Take 1.5-5ml clone E40-6B bacteria solution, centrifuge at 13,000rpm for 3min, discard the supernatant, collect the bacteria, and suck up the supernatant as much as possible;
[0038] (2) Add 200 μl of RNase A-added buffer T1 to the cells obtained in step (1) to resuspend the cells, shake and mix to obtain a resuspension;
[0039] (3) Add 200 μl of buffer T2 to the resuspension prepared in step (2), mix by gently inverting 5-10 times to obtain a clear lysate, and place at room temperature for 5 minutes;
[0040...
Embodiment 3
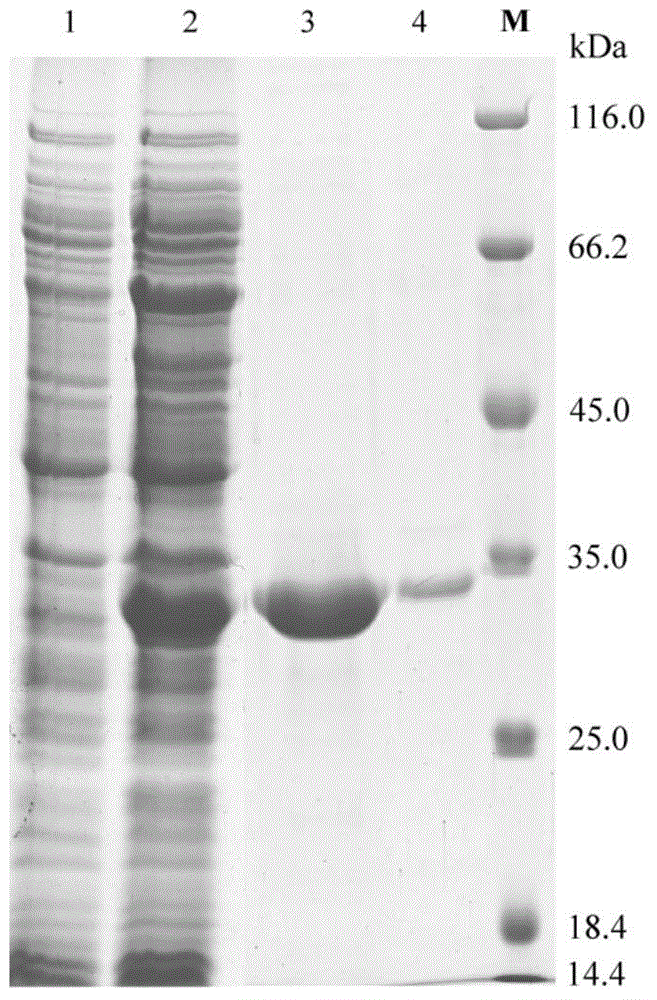
[0053] Example 3: Cloning, heterologous expression and separation and purification of esterase E40
[0054] 2.1 Amplification of E40 gene sequence by PCR
[0055] (1) Design two specific primers according to the E40 gene sequence:
[0056] 40F: CGG CATATG GCCAAAAGCCCAGAGTT (SEQ ID NO.3), underlined is the NdeI restriction site;
[0057] 40R: GCC AAGCTT TCAGCCGATCTGCTTCCGC (SEQ ID NO.4), underlined is the HindIII restriction site;
[0058] Primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd.
[0059] (2) Using 40F and 40R as primers and using the fosmid where E40 is located as a template, FastPfu DNA polymerase (purchased from Transgen) was used to amplify the target gene fragment.
[0060] The PCR reaction conditions were: pre-denaturation at 95°C for 2 min; then denaturation at 95°C for 20 sec; annealing at 55°C for 20 sec; extension at 72°C for 20 sec, after 30 cycles; extension at 72°C for 10 min.
[0061] (3) PCR amplification product is carried out ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com