Gene engineering strain capable of safely and efficiently producing Phenazino-1-carboxylic acid and application thereof
A technology of genetically engineered strains and sulfazinamycin, applied in microorganism-based methods, bacteria, microorganisms, etc., can solve the problems of low fermentation titer, increased metabolic flux, unfavorable for industrial production and large-scale promotion and use, etc. To achieve the effect of superior safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
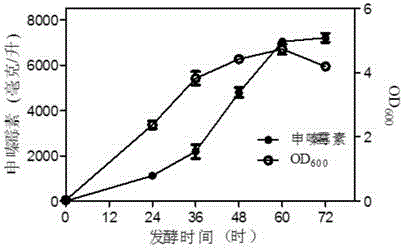
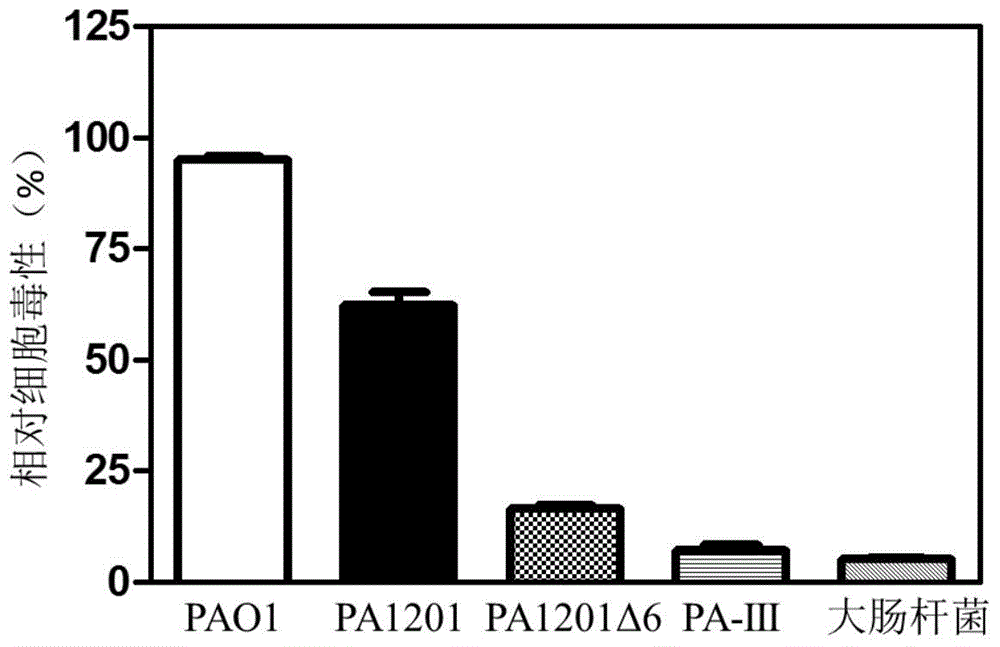
Image
Examples
Embodiment 1
[0125] This embodiment involves knocking out the pabB gene, comprising the following steps:
[0126] 1. Construction of recombinant suicide plasmid pEX-pabB:
[0127] 1.1 Design two pairs of primers (pabB-FOR-1 and pabB-REV-1, pabB-FOR-2 and pabB-REV-2) to amplify the upstream and downstream flanking sequences of the gene pabB respectively. The nucleotide sequences of the primers are shown in the table 2 shown in SEQ ID No.37 and SEQ ID No.38, SEQ ID No.39 and SEQ ID No.40.
[0128] 1.2 Using the genomic DNA of strain PA1201 as a template, use the high-fidelity polymerase KOD-plus-neo and the designed primers to amplify the two flanking sequences of the gene pabB. The PCR product is detected by 0.8% agarose gel electrophoresis and gelled by AxyPrep Gel Recovery Kit recovered two flanking fragments, the lengths were 466bp and 907bp ( Figure 5 ).
[0129] 1.3 Using the above two flanking fragments as templates, using pabB-FOR-1 and pabB-REV-2 as primers, use the high-fidelit...
Embodiment 2
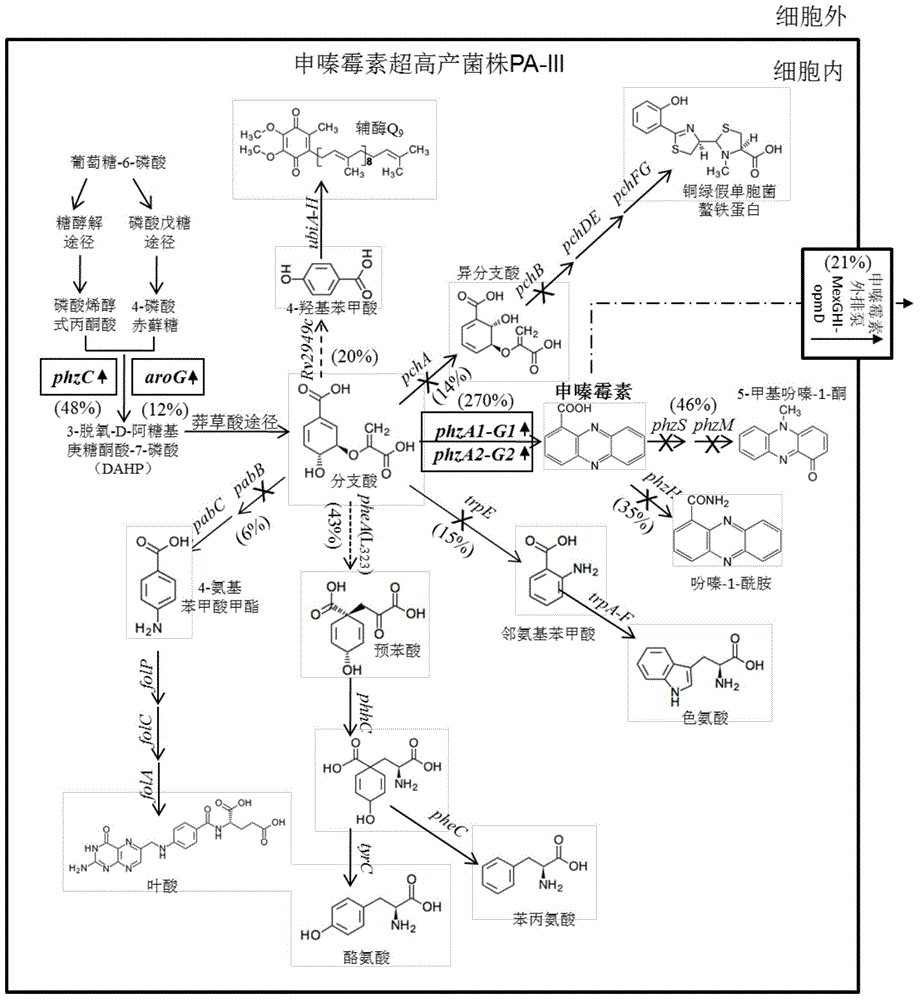
[0142] In this example, by constructing the genome integration vector mini-Tn7T-Gm-phzC, inserting the full gene sequence of phzC at the attTn7 site of the genome of PA1201 and its derivative engineering strains to increase the expression copy number and expression level of phzC ( Figure 8 ), including the following steps:
[0143] 1. Construction of genome integration vector mini-Tn7T-Gm-phzC
[0144] 1.1 A pair of primers (iphzC-FOR and iphzC-REV) were designed to amplify the coding sequence of the phzC gene using the high-fidelity polymerase KOD-plus-neo using Pseudomonas aeruginosa PA1201 genomic DNA as a template. The obtained PCR product was detected by 0.8% agarose gel electrophoresis, and the fragment was recovered by using the AxyPrep gel recovery kit, and the length was 1640bp. The nucleotide sequences of the primers are shown in Table 2 as SEQ ID No.53 and SEQ ID No.54.
[0145] 1.2 Design a pair of primers (iptac-FOR and iptac-REV), use the pME6032 plasmid DNA a...
Embodiment 3
[0160] In this example, the PA1201 derived engineering strain PA1201MSHΔ 9pheA (W323L) Rv2949c (P tac -phzC) the aroG gene promoter on the genome was replaced by P tac The method for a strong promoter specifically comprises the following steps:
[0161] 1. Construction of recombinant suicide plasmid pEX-P tac -aroG
[0162] 1.1 Design 2 pairs of primers (r2020-FOR-1, r2020-REV-1 and raroG-FOR-3, raroG-REV-3), using the genomic DNA of Pseudomonas aeruginosa PA1201 as a template, using high-fidelity The polymerase KOD-plus-neo amplified the upstream flanking sequences of aroG and aroG, the PCR products were detected by 0.8% agarose gel electrophoresis, and the two flanking fragments were recovered by AxyPrep Gel Recovery Kit, the lengths were 680bp and 640bp, respectively. The nucleotide sequences of the primers are shown in Table 2 as SEQ ID No.61, SEQ ID No.62, SEQ ID No.63 and SEQ ID No.64.
[0163] 1.2 Design a pair of primers (rPtac-FOR-2 and rPtac-REV-2), use the pME60...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com