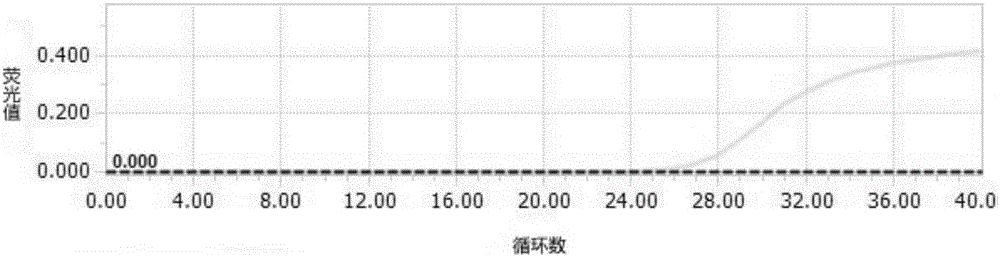
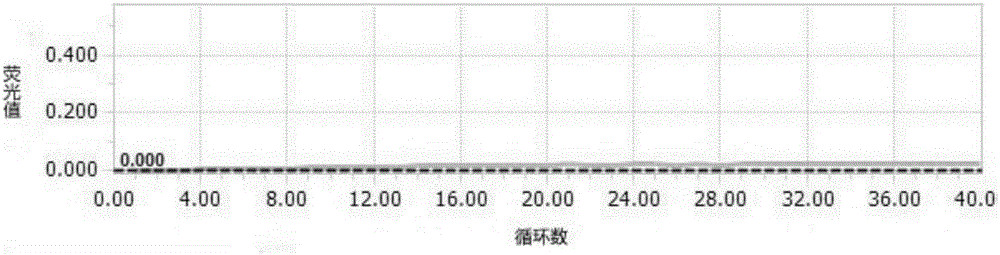
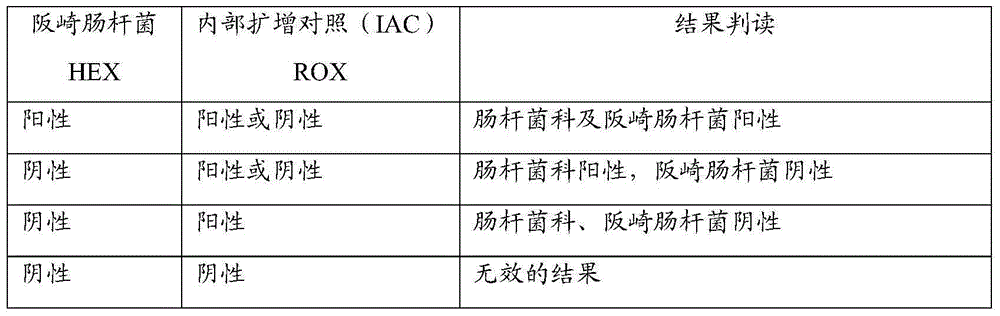
Primer and probe for specific detection on enterobacter sakazakii and applications of primer and probe
An Enterobacter sakazakii, specific technology, applied in the field of microbial detection, can solve the problems of insufficient rigor and accuracy, large medium raw materials, difficult to determine results, etc., to save laboratory space, ensure accuracy, and avoid false negatives. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] In this example, specific kit products are used to detect Enterobacter sakazakii in food, especially in infant formula milk powder. The kit product is based on the method of real-time quantitative PCR and includes the following reagents:
[0095] Reagent S: ethidium bromide monoazide bromine aqueous solution 125 μg / mL;
[0096] Reagent L: DNA extraction solution, namely 0.1% Chelex100 (chelating resin) aqueous solution, can extract the DNA of Gram-negative bacteria (such as Enterobacteriaceae) within 30 minutes, and the extracted DNA can be directly used as a DNA template for PCR ;
[0097] Reagent 1: Contains specific primers and probes, the final concentration of the primers is 0.4 μmol / L, the final concentration of the probes is 0.2 μmol / L, MgCl 2 , dNTPs, PCR reaction buffer;
[0098] The sequence structure of the primers is as follows:
[0099] Enterobacter sakazakii-F: 5′-AGGGGATATTGTCCCCTGAAACAG-3′;
[0100] Enterobacter sakazakii-R: 5′-AAACGAGAATAAGCCGCGCAT...
Embodiment 2
[0138] In this example, product A is used to detect Enterobacter sakazakii and other Enterobacteriaceae bacteria in food, especially in infant formula milk powder. A product is based on the method of real-time quantitative PCR, including the following reagents:
[0139] Reagent S: ethidium bromide monoazide bromine aqueous solution 125 μg / mL;
[0140] Reagent L: DNA extraction solution, namely 0.1% Chelex100 (chelating resin) aqueous solution, can extract the DNA of Gram-negative bacteria (such as Enterobacteriaceae) within 30 minutes, and the extracted DNA can be directly used as a DNA template for PCR ;
[0141] Reagent 1: Contains specific primers and probes, the final concentration of the primers is 0.4 μmol / L, the final concentration of the probes is 0.2 μmol / L, MgCl 2 , dNTPs, PCR reaction buffer;
[0142] The sequence structure of the primers is as follows:
[0143] Enterobacter sakazakii-F: 5′-AGGGGATATTGTCCCCTGAAACAG-3′;
[0144] Enterobacter sakazakii-R: 5′-AAAC...
Embodiment 3
[0186] The method for the simultaneous specific detection of Enterobacteriaceae bacteria and Enterobacter sakazakii described in this example uses the kit described in Example 2, specifically comprising:
[0187] (1) Extraction of DNA (using reagent S and reagent L)
[0188] Take 100 g (mL) of the test sample and dissolve it in an Erlenmeyer flask filled with 900 mL of buffered peptone water preheated to 37 °C, and incubate at 37 °C for 18 hours for the first enrichment. Pipette 450 μl of buffered peptone water into a 1.5 ml centrifuge tube. Add 50 μl of the first enriched sample and mix well. Incubate at 37°C for 3-4h for secondary enrichment.
[0189] Pipette 300 μl of Reagent S (ethidium bromide monoazide bromide (EMA): 125 μg / mL) into a new 1.5ml centrifuge tube, then add 100 μl of the secondary enriched sample, and then incubate in the dark room for 5 minutes, then incubate under 500W light Lower exposure for 5min. Centrifuge at 8,000 g for 5 min and remove the supern...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com