Vitreoscilla hemoglobin mutant and controllable expression of vitreoscilla hemoglobin mutant in genetically engineered bacteria
A technology for hemoglobin and Vibrio vitreous bacteria, which is applied in genetic engineering, hemoglobin/myoglobin, plant genetic improvement, etc., can solve the problems of difficulty in meeting oxygen, complex α-amylase synthesis pathway, and consumption of a large amount of oxygen, so as to improve utilization efficiency, increase output, and reduce material loss
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
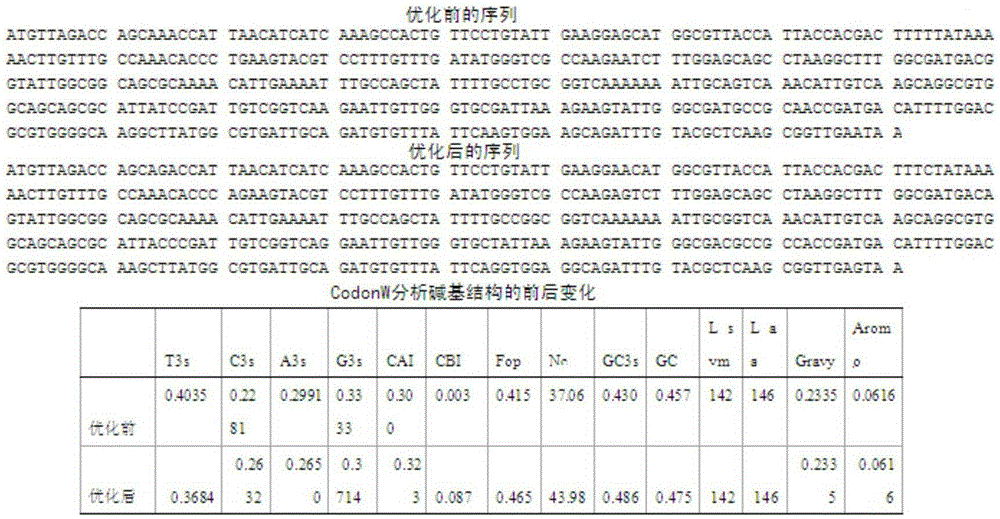
[0039] Codon optimization and site-directed mutagenesis were performed on the hemoglobin gene sequence of Vitiligo hyaline to obtain the sequence.
[0040] According to the Vibrella hyaline hemoglobin gene sequence (as shown in SEQIDNO: 1) and Bacillus subtilis codon preference in GenBank, use software CodonW software package to analyze gene structure and site-directed mutagenesis kit, select certain sites to carry out site-directed mutagenesis and After codon optimization, the gene sequence was preliminarily determined by analyzing CAI, CBI, and Nc values. Analyze the mRNA free energy by RNAstructure, combine the preferred codons of Bacillus subtilis, and finally select the modification scheme with lower free energy, and design the codons with the preferred codons of Bacillus subtilis after adding the restriction sites of BamH1 and Sal1 at both ends of the sequence The hemoglobin mutant gene sequence of Vitiligo hyaline (as shown in SEQ ID NO: 2), and then this gene sequence ...
Embodiment 2
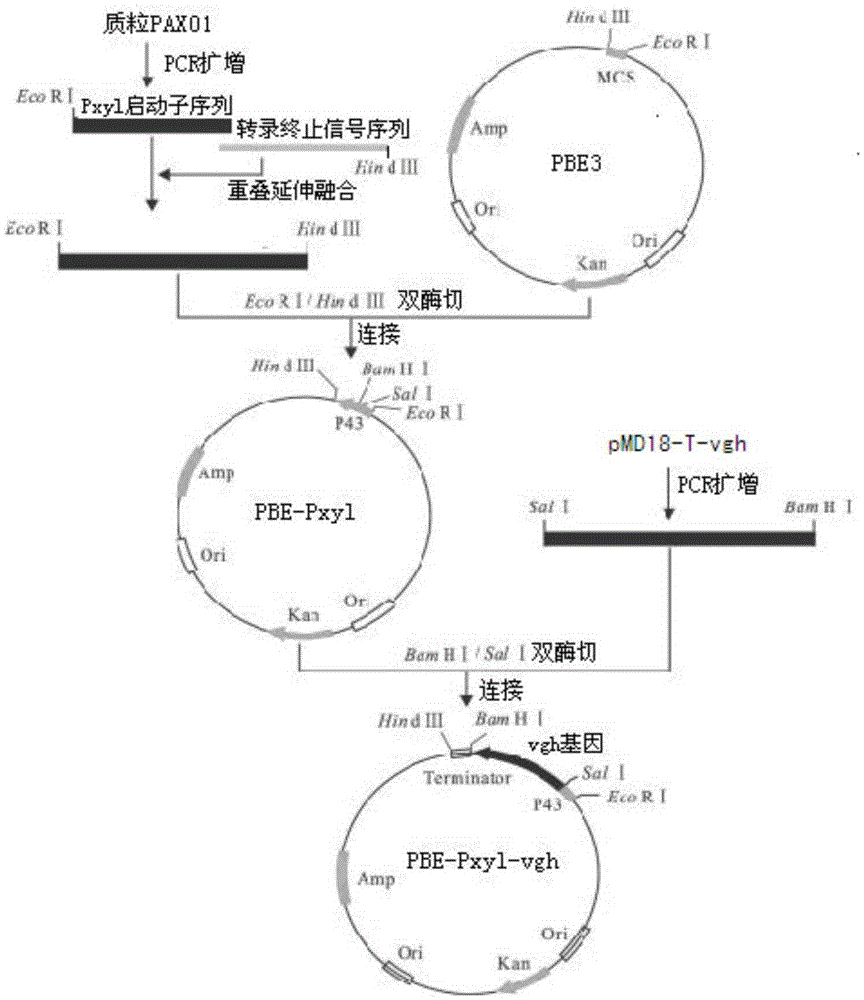
[0042] Construction of the hemoglobin gene expression vector PBE-Pxyl–vgh containing codon-optimized and site-directed mutations and the acquisition of recombinant strains (Bacillus subtilis / PBE-Pxyl-vgb).
[0043] Build process like figure 2 As shown, the xylose promoter Pxyl fragment on the plasmid pAX01 was amplified with primers Y1 (containing the EcoR1 restriction site) and Y2, and the primer sequences were as follows:
[0044] Y1: CCGGAATTCAACCATTTGCTGTTGCTTGA (SEQ ID NO: 5),
[0045] Y2: CGCCAATGCTTGTCACGAATATAAGATA (SEQ ID NO: 6);
[0046] The PCR reaction system and procedures are as follows:
[0047]
[0048] The transcription termination signal sequence on the plasmid pAX01 was amplified with primers Y3 and Y4 (containing HindIII restriction sites), and the primer sequences were as follows:
[0049] Y3: CTTATATTCGTGACAAGCATTGGCGATCGGCTGTTTGG (SEQ ID NO: 7),
[0050] Y4: CCCAAGCTTTCCCCCTACTGCGTGTCGTA (SEQ ID NO: 8);
[0051] The PCR reaction system and proce...
Embodiment 3
[0081] The expression of hemoglobin was detected by carbon monoxide differential light spectroscopy.
[0082]After culturing the original bacteria and the recombinant bacillus overnight, inoculate them in 60ml of fermentation medium with 1% by volume, and culture them with shaking at 37°C for 12h (wherein 3% xylose is added at 7h to induce codon-optimized Vibrella hyaline The expression of bacterium), take 15mL bacterium liquid, 8000r / min, after 10min centrifugation collects thalli, after washing three times with 50mM, the potassium phosphate buffer of pH7.0, resuspend in 10mL potassium phosphate buffer. Under the condition of ice bath, the bacterial suspension was ultrasonically oscillated with an action time of 25s and an interval of 50s to break up the cells. Add 20 mg of sodium dithionite per milliliter to the bacterial suspension of broken cells, and then divide it into 2 parts of equal volume. One part is fed with carbon monoxide gas for 5 minutes as a sample, and the ot...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com