Specific probe and method thereof for rapid and sensitive quantification of staphylococcus aureus
A staphylococcus, golden yellow technology, applied in the intersection of microbiology and molecular biology, can solve the problem of the research lag of pathogenic bacteria detection method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Specificity Verification of Nucleic Acid Probes
[0027] 1.1 Strains
[0028] Escherichia coli, Shigella flexneri, Shigella sonii, Salmonella typhimurium, Salmonella choleraesuis, Proteus vulgaris, Bacillus subtilis, Bacillus licheniformis, Staphylococcus aureus, Aeromonas hydrophila Enterococcus faecium
[0029] 1.2 Oligonucleotide probes
[0030] The test uses Staphylococcus aureus specific probe SA-1, the sequence is: 5'-GCCGGTGGAGTAACCTTTTAGGAGC-3'. Among them, the 5' end was labeled with FITC by fluorescein isothiocyanate, and the probe was synthesized and labeled by Dalian Bao Biological Company.
[0031] 1.3 FISH-FCM method to verify probe specificity
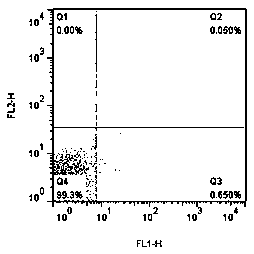
[0032] Obtain a single colony of eleven kinds of bacterial strains according to the conventional planning line culture method, cultivate them in liquid medium, and adjust the bacterial concentration to 10 6cells / ml, and the hybridization method is as follows: (1) Take 200 μl of bacterial liquid and fix it wit...
Embodiment 2
[0039] Sensitivity of FISH-FCM to detect the content of Staphylococcus aureus
[0040] 2.1 Nucleic acid probe
[0041] Ditto.
[0042] 2.2 Detection of samples by FISH-FCM method
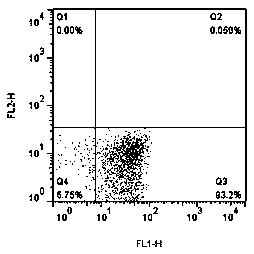
[0043] Counted under a microscope, the original concentration of Staphylococcus aureus liquid was 5.0×10 6 pieces / ml. 10 times the ratio of diluted bacterial solution to 5.0×10 6 , 5.0×10 5 , 5.0×10 4 , 5.0×10 3 , 5.0×10 2 , 5.0×10 1 Gradient bacterial solution was tested. The detection method is as above 1.3 (1)-(4), adding 50 μl of 1000 / μl fluorescent microspheres (CountBringhtTM, Invitrogeon, USA) to the mixture, and analyzing 5000 objects for each sample by flow cytometry.
[0044] 2.3 Result Analysis
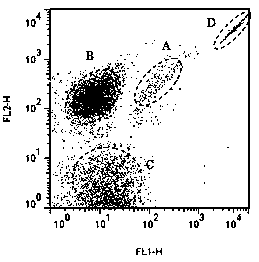
[0045] Analysis was performed with CellQuestPro software (version 5.2.1, Becton Dickinson, USA). On the dotplot graph, the X-axis represents the fluorescence intensity of the target bacteria hybridized with the fluorescent probe, and the Y-axis represents the fluorescence intensity of...
Embodiment 3
[0054] Comparison of FISH-FCM method and traditional culture method in the detection of Staphylococcus aureus in meat samples
[0055] 3.1 Nucleic acid probe
[0056] Ditto.
[0057] 3.2 Detection of samples by FISH-FCM method
[0058] The pretreatment of fresh meat samples is the same as that of SNT0738-1997 to prepare 10 -1 For meat sample liquid, take 1ml of meat sample liquid and centrifuge at 80g for 1min at low speed to remove impurities. The remaining hybridization methods are the same as above, prepare the liquid in step (5) above, add 10 μl propidium iodide (0.5 mg / ml), and incubate at 37°C in the dark for 20 minutes. Add 1000 / μl fluorescent microspheres (CountBringhtTM, Invitrogeon, USA) to 50 μl. The mixed solution was loaded on the flow cytometer for analysis. 10,000 objects were analyzed per sample.
[0059] 3.3 Samples detected by culture method
[0060] According to the national standard GB / T4789.10-2010 method.
[0061] 3.4 Result analysis
[0062] Dit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com