Sequential enzyme surface co-display system and use thereof
A surface co-display and co-display technology, applied in the field of biotechnology and analysis, can solve the problems of reduced detection efficiency, loss of enzyme activity, low efficiency, etc., to solve the complex extraction process, low stability, and good stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Acquisition of cell surface display sequence enzyme expression vector:
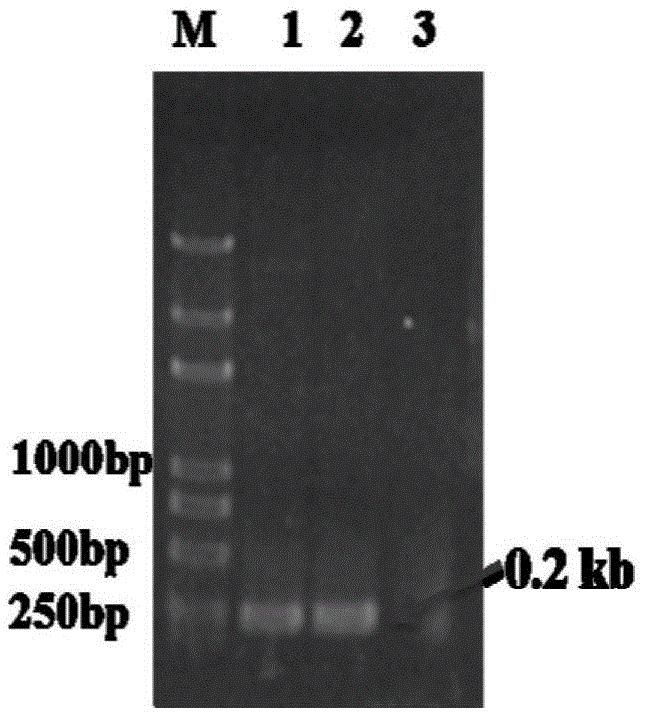
[0047] 1. The gene fragment docc encoding the dockerin protein comes from the strain Clostridium cellulolyticumH10; doct comes from the strain ClostridiumthermocellumATCC27405, wherein the length of the docc gene fragment is 213bp, and the length of the doct gene fragment is 204bp. According to the gene sequences of the two, primers are used from the respective genomic DNAs with high-fidelity polymerases Amplify the gene of interest separately. The target gene PCR reaction system is as follows: 0.25μL ExTaq, 5μL 10×ExTaqbuffer, 4μL 2.5mMdNTP, 1μL genomic DNA, 1μL DocC-F and DocT-F, 1μL DocC-R and DocT-R, 37.75μL ultrapure water. PCR reaction conditions: 94°C for 5min; 94°C for 30s, 58°C for 30s, 72°C for 20s, 30 cycles; 72°C for 5min; after gene amplification, use 1% agarose gel to detect the size and purity of the target gene (see figure 1 ), and then ligated to the pMD-19Tsimple vector (TaKaRa, ...
Embodiment 2
[0059] Acquisition of cells displaying sequence enzymes on the cell surface:
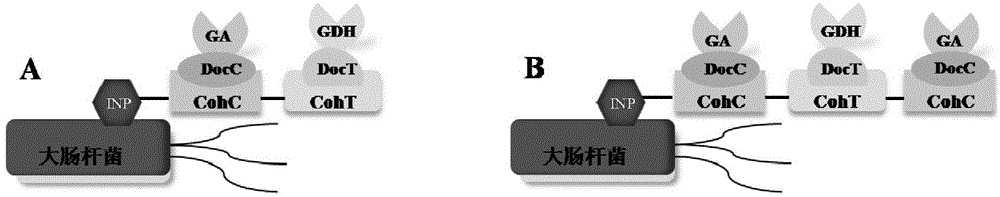
[0060] 1. Expression of fusion protein. The recombinant vectors obtained in Example 1 were respectively transformed into Escherichia coli Escherichiacoli BL21 (DE3), inoculated in fresh LB liquid medium containing kanamycin (30 μg / ml) after enzyme digestion and identification, and cultured with shaking until the absorbance (OD600 ) to 0.6, add 0.5mM, 0.2mM, and 1mM IPTG (isopropyl-β-D-thiogalactopyranoside) respectively, culture at 25°C for 20h, and induce the expression of GA-DocC and DocT-GDH respectively , INP-CohC / CohT and INP-CohC / CohT / CohC fusion proteins.
[0061] Escherichia coli E.coliBL21 (DE3) was cultured in LB medium, and the composition of LB medium was: 5g / L yeast extract, 10g / L peptone, 10g / L NaCl.
[0062] 2. Dispense the bacteria liquid into 50mL centrifuge tubes and centrifuge at 6000rpm for 5min, discard the supernatant, wash twice with PBS buffer of pH 7.4 after collecting all...
Embodiment 3
[0066] Determination of single-enzyme assembler activity of GA-DocC and DocT-GDH:
[0067] 1. Enzyme activity assay of GA-DocC single enzyme assembly. Take 2.5ml GA-DocC crude enzyme solution and mix with 20μl INP-CohC / CohT / CohC bacterial solution. Incubate at 25°C and 180rpm for 2h, centrifuge at 6000rpm for 3min to remove the supernatant, wash twice with the same buffer solution, and measure GA enzyme activity. With 10mM maltose as substrate, react at 70°C for 15min. After the reaction was completed, the glucose concentration in the product was measured with a glucose kit. Finally, the enzymatic activity of GA-DocC single-enzyme assembly was calculated. The GA activity unit U is defined as the amount of enzyme required to release 1 μmol of glucose per minute under certain reaction conditions. The enzyme activity of whole cells displayed by GA at a ratio of 2:1 is 0.82U / OD 600 whole cells.
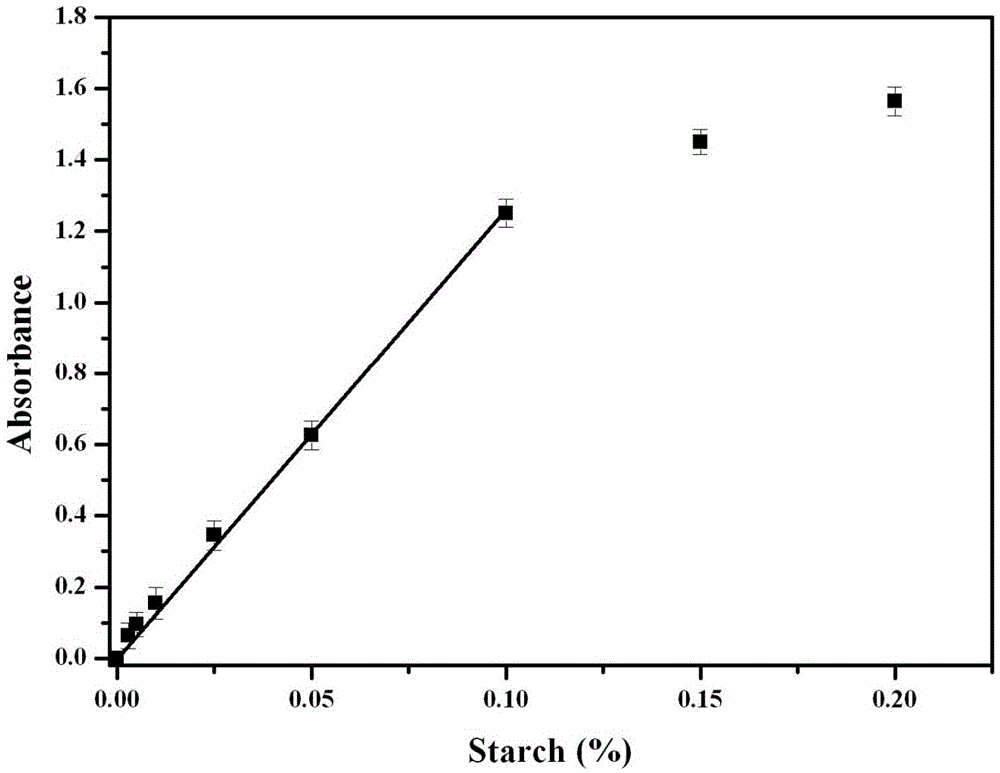
[0068] 2. NADH standard curve preparation. NADH is composed of NAD + It is re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com