Method for detecting yeast beta-glucan in milk or milk product
A detection method and technology for dairy products, applied in the field of analysis and detection, can solve problems such as the lack of detection standards and related technologies, affecting the extraction and purification of yeast β-glucan, and the complexity of the dairy matrix
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
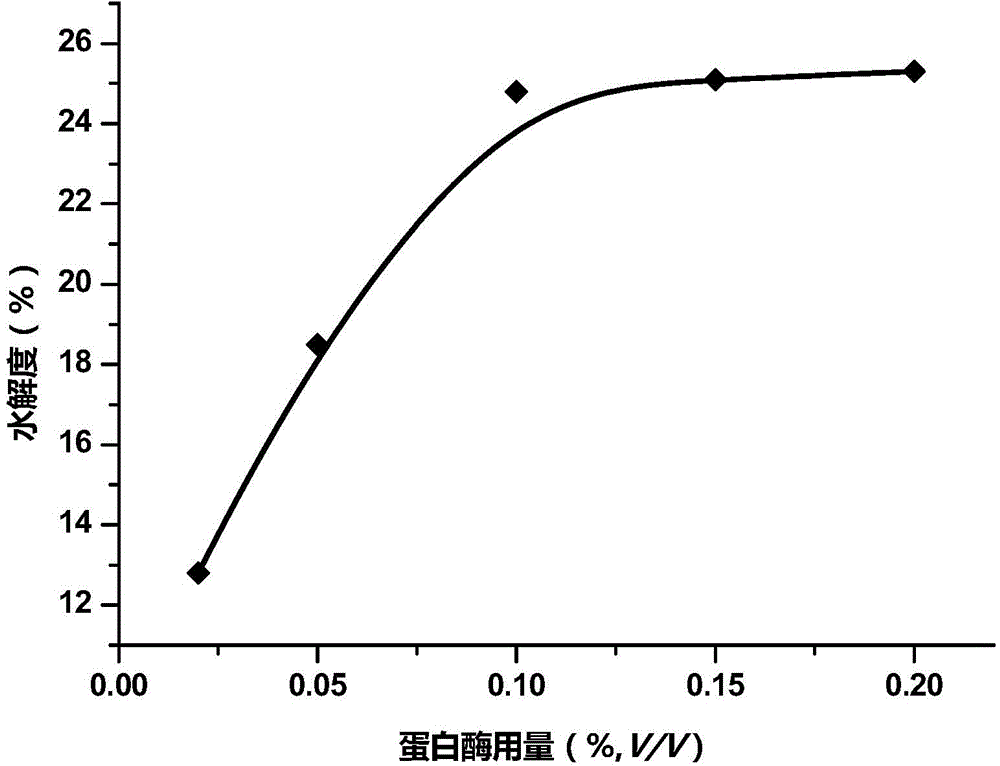
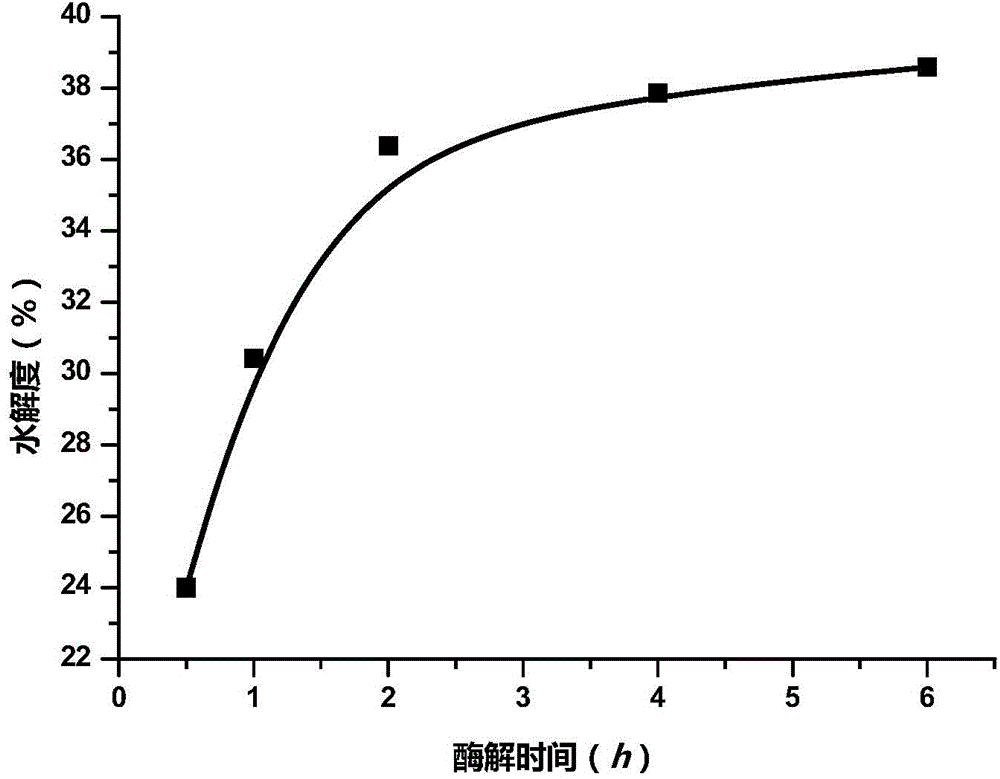
[0051] Embodiment 1: the relation of the dosage of neutral protease and alkaline protease, enzymolysis time and sample proteolysis degree
[0052] In this example, the degree of proteolysis is used as an index to measure the degree of proteolysis.
[0053] 0.02%, 0.05%, 0.1%, 0.15% and 0.2% (v / v) by volume to milk supplemented with yeast β-glucan neutral protease stock solution and Equal-volume mixed solution of alkaline protease stock solution (converted according to the density mentioned above, in the milk to be enzymatically hydrolyzed, the enzyme activity of Neutrase neutral protease is 1.0×10 -4 AU / mL, 2.5×10 -4 AU / mL, 5.0×10 -4 AU / mL, 7.5×10 -4 AU / mL, 1.0×10 -3 AU / mL, the enzyme activity of Alcalase alkaline protease is 2.8×10 -4 AU / mL, 7.0×10 -4 AU / mL, 1.4×10 -3 AU / mL, 2.1×10 -3 AU / mL, 2.8×10 -3 AU / mL), the enzymatic hydrolysis was carried out at 45°C for 6 hours. After the enzymatic hydrolysis was completed, the degree of proteolysis of each sample was det...
Embodiment 2
[0055] Embodiment 2: the mensuration of actual sample
[0056] Samples 1-12 (see Table 1 for sample numbers) are all milk samples added with 42 mg / 100 g of yeast β-glucan.
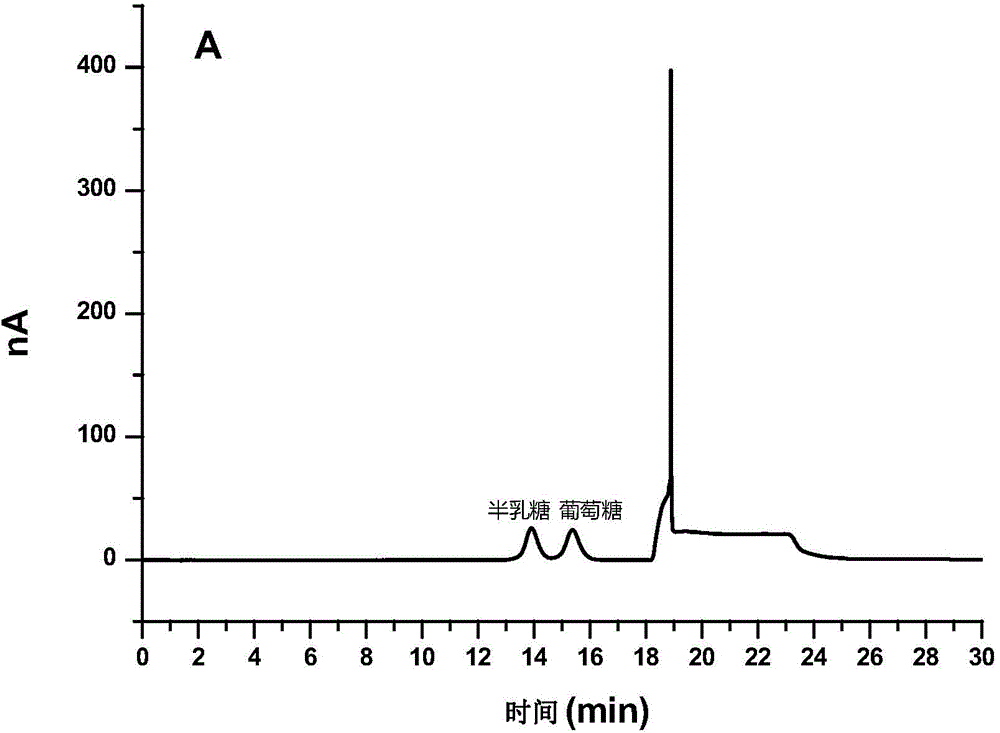
[0057] 1) Pretreatment step: add 0.1% (v / v) to 12 yeast β-glucan milk samples respectively neutral protease stock solution and Equal volumes of the alkaline protease stock solution were hydrolyzed at 45° C. for 2 hours, and the obtained hydrolyzed product was centrifuged at 10,000 rpm. The supernatant was discarded, and the obtained crude dextran precipitate was washed twice with pure water. Then add 1 mol / L hydrochloric acid to the precipitate and hydrolyze at 121°C for 1 hour.
[0058] 2) Analysis step: the glucose in the sample after the pretreatment step is determined by ion chromatography. Before the ion chromatographic analysis, the dextran hydrolysis solution obtained from the pretreatment was diluted by direct dilution method, so that the concentration of glucose to be determined was in the l...
Embodiment 3
[0076] Embodiment 3: Sensitivity, accuracy and precision analysis of the method
[0077] Use pure water to prepare glucose standard solutions and sample solutions with a series concentration of 0.5, 1.0, 4.0, 8.0, 10.0, 15.0, and 20.0 μg / mL, and evaluate the sensitivity, accuracy, and precision of the analysis.
[0078] Under the same chromatographic conditions as in Example 2, the linear regression equation obtained by measuring the glucose standard solution is Y=2.865X+0.395, the linear range is 0.5~20 μg / mL, the linear correlation coefficient is 0.9995, and the detection limit is 0.1 μg / mL. The relative standard deviation was 0.15% (n=6).
[0079] Taking the concentration of yeast β-glucan added in dairy products at 42.00mg / 100g as the standard sample, prepare samples with three addition levels of 70%, 100% and 130% of the standard sample, and determine the sample recovery rate at the three concentrations : when the addition amount was 70%, the measured value was 22.32mg / 1...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com