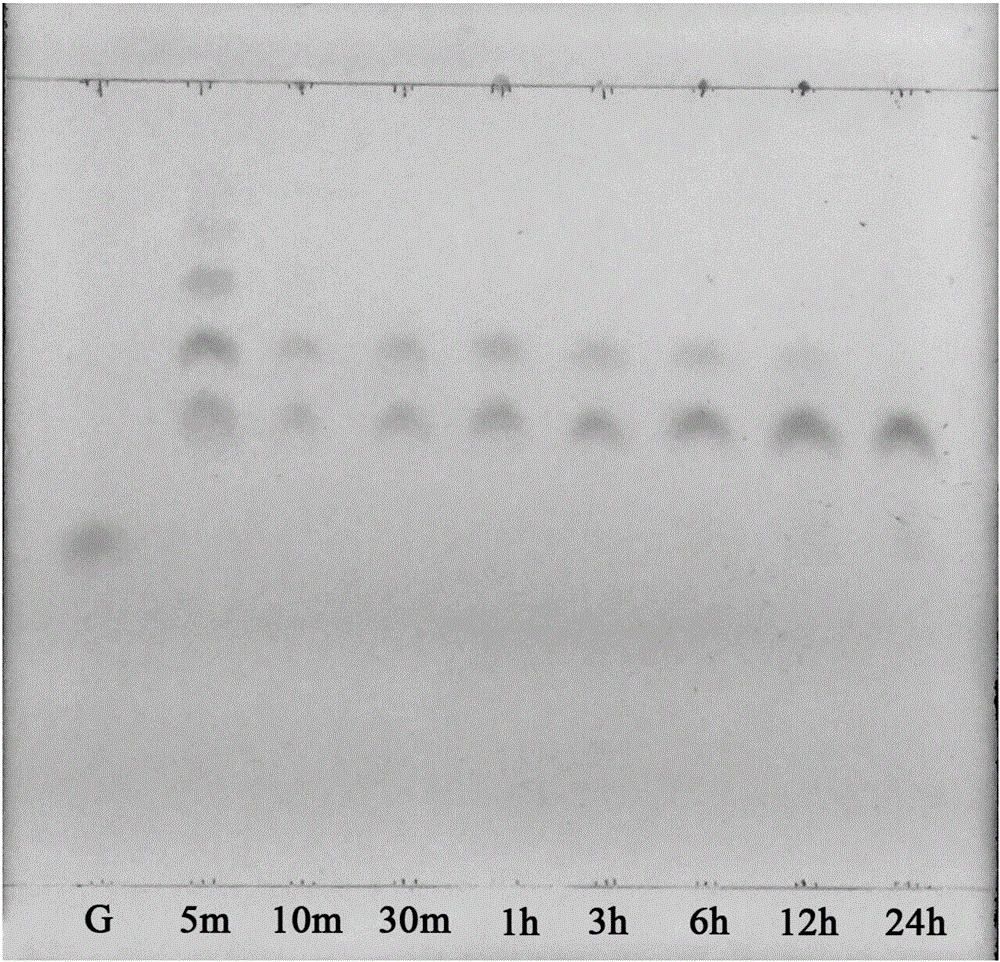
Beta-agarase and coding gene and application thereof
A kind of agarase and gene technology, which is applied in application, genetic engineering, plant gene improvement, etc., can solve the problems of difficult large-scale production, instability, low activity, etc., and achieve easy purification, good enzyme activity, and good thermal stability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Example 1: Acquisition of β-agarase gene Aga0917
[0022] The entire genome analysis of the agarase-producing Pseudoalteromonas citrea (Pseudoalteromonas citrea) YTW1-15-1 strain was carried out, and the upstream and downstream primers were designed as follows:
[0023] Upstream primer: 5'-CGCGGATCCGCAGATTGGGACGCATATAG-3',
[0024] Downstream primer: 5'-CCGCTCGAGTTAGTTTGCTTTGTAAACACG-3'.
[0025] The Pseudoalteromonas citrea YTW1-15-1 bacterial strain was inoculated in the marine broth 2216 liquid medium, shaken and cultivated at 28°C and 120rpm for 24h, took 1.5ml of the culture solution, and followed the method in the instruction manual of the bacterial genomic DNA extraction kit (Takara), Genomic DNA of Pseudoalteromonas citrea YTW1-15-1 strain was extracted.
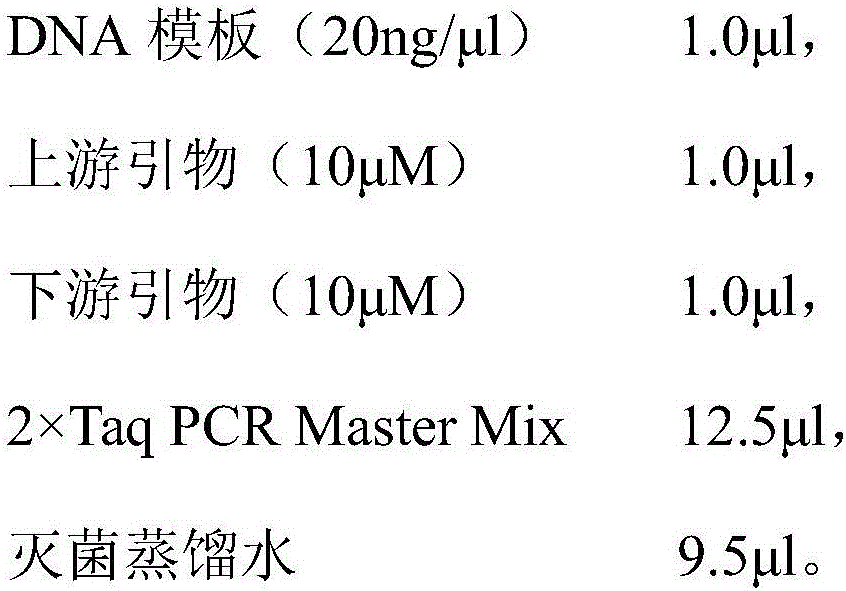
[0026] Using the genomic DNA of Pseudoalteromonas citrea YTW1-15-1 strain as a template, the PCR reaction was carried out under the action of the above-mentioned upstream and downstream primers. After the P...
Embodiment 2
[0031] Example 2: Construction of β-agarase gene cloning vector pMD19T-Aga0917
[0032] In order to increase the concentration of the target gene fragment (β-agarase gene Aga0917) when it is connected with the expression vector and ensure the complete digestion of the target gene fragment and reduce the false positive results of the recombinant vector, this example uses pMD19T vector and β-agar The enzyme gene Aga0917 was used to construct the cloning vector pMD19T-Aga0917.
[0033] The purified β-agarase gene Aga0917 was ligated with the pMD19T vector using a DNA ligation kit (DNA Ligation Kit) to obtain the ligation product—β-agarase gene cloning vector pMD19T-Aga0917.
[0034] The ligation reaction system is as follows (10 μl):
[0035] pMD19T vector (50ng / μl) 1.0μl,
[0036] β-agarase gene Aga0917 (165ng / μl) 4.0μl,
[0037] DNA ligation kit 5.0 μl.
[0038] Ligation reaction conditions: Incubate the reaction at 16°C for 16 hours.
Embodiment 3
[0039] Example 3: Construction of Escherichia coli recombinant strain DH5α-pMD19T-Aga0917
[0040] Thaw 100 μl on ice to a concentration of 5 x 10 7 cfu / ml Competent E. coli DH5α cells and 10 μl of the ligation product obtained in Example 2 were mixed, ice-bathed for 30 minutes, heat-shocked at 42°C for 90 seconds, quickly placed in ice for 3 minutes, and 890 μl of LB liquid medium was added. Recover on a shaker at 37°C and 170r / min for 1h, then centrifuge at 4°C and 4000r / min for 5min, suck out 890μl of supernatant, mix the remaining supernatant with bacteria, and mix the bacteria with 8μl of isopropyl - β-D-thiogalactopyranoside (IPTG), 40μl 5-bromo-4-chloro-3-indole-β-Dgalactoside (X-gal) was mixed and spread evenly in the solution containing 100μg / ml ampicillin On the LB solid medium of penicillin, place it upright for about 10 minutes, let the surface of the plate dry, then invert it, and incubate at 37°C for 12-16 hours.
[0041] White colonies were picked, and their p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com