Biosynthetic gene cluster of noveltymycin and its application
A biosynthetic and noveltymycin technology, applied in the field of microbial genetic resources and genetic engineering, can solve undiscovered problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
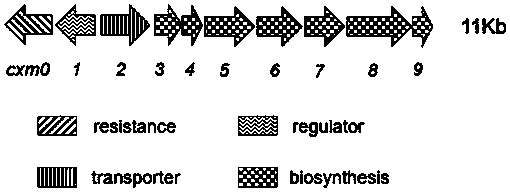
[0049] Example 1: Extraction and sequencing of the genome DNA of Actinomycetes florensis Jinan, a noveltymycin-producing bacterium
[0050] Inoculate the Jinan Actinomycetes strain from -80°C to a Gowell solid plate, culture at 30°C for 4 days, scrape the bacteria and inoculate to 168G (glucose 22g, beef extract 4g, peptone 5g, yeast extract 0.5g, casein Add 3g of peptone, 1.5g of table salt, add 900mL of water to sterilize at high temperature, add 10g of glycine to 100mL of water to sterilize separately, add 1 / 10th volume of 10% glycine to the medium when in use) medium 50mL / 250mL Erlenmeyer flask, 30℃ , cultivated at 150rpm for 4 days, the cells formed milky white pellets, put 1.8mL into 2mL centrifuge tubes, centrifuged at 13000rpm for 5 minutes, poured off the supernatant and washed with 1.6mL of water, centrifuged at 13000rpm for 5 minutes, discarded the supernatant, Resuspend the cells with 450μL of 10mM Tris-HCl pH8.0, add 30μL of 20mg / mL proteinase K, mix well, add 40μ...
Embodiment 2
[0055] Example 2: Direct cloning of noveltymycin biosynthetic gene cluster
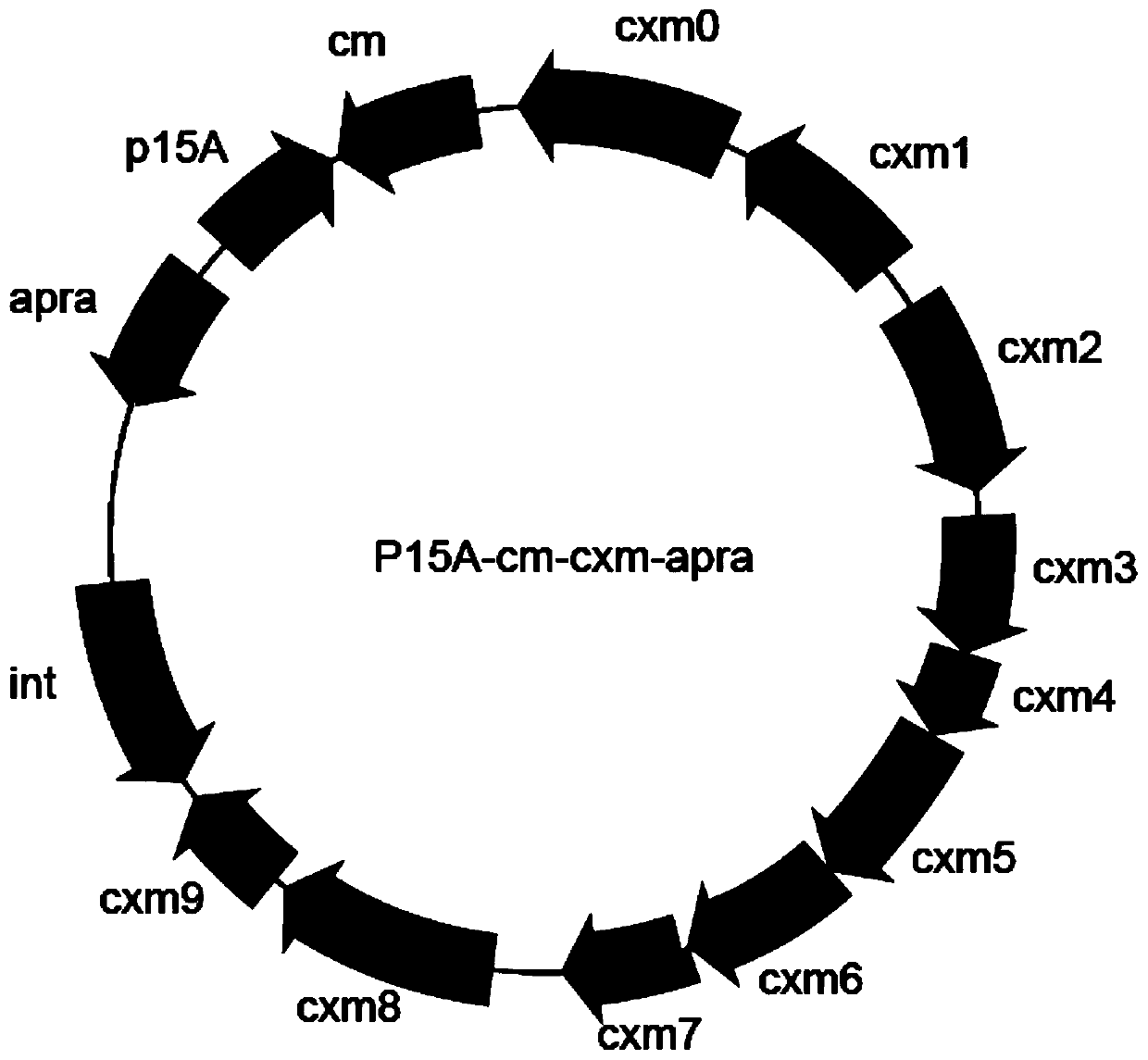
[0056] After Genomic DNA was extracted from Actinomycetes jinanensis, take 200 μl, about 300 μg / μl, and perform double enzyme digestion with EcoRV and AscⅠ, 400 μl enzyme digestion system, after incubating at 37°C for 3 hours, extract DNA once with phenol chloroform, then ethanol Precipitation recovery, with a small amount of ddH 2 O dissolved. The linear cloning vector p15A-cm-HA, HA is the homologous arm of the cenomycin gene cluster, obtained by amplification of the plasmid p15A-cm-tetR-ccdB-hgy. 5 μg of genomic DNA and 0.5 μg of linear cloning vector were co-electroporated into GB05dir expressed by induced recombinase, recovered at 37°C for 1 hour, spread to LB plates containing cm, cultured overnight at 37°C, and clones were picked with a medium containing cm LB was cultured overnight, the plasmid was extracted and identified by enzyme digestion, the recombinant plasmid p15A-cm-cxm containing t...
Embodiment 3
[0057] Example 3: Heterologous expression of noveltymycin biosynthetic gene cluster
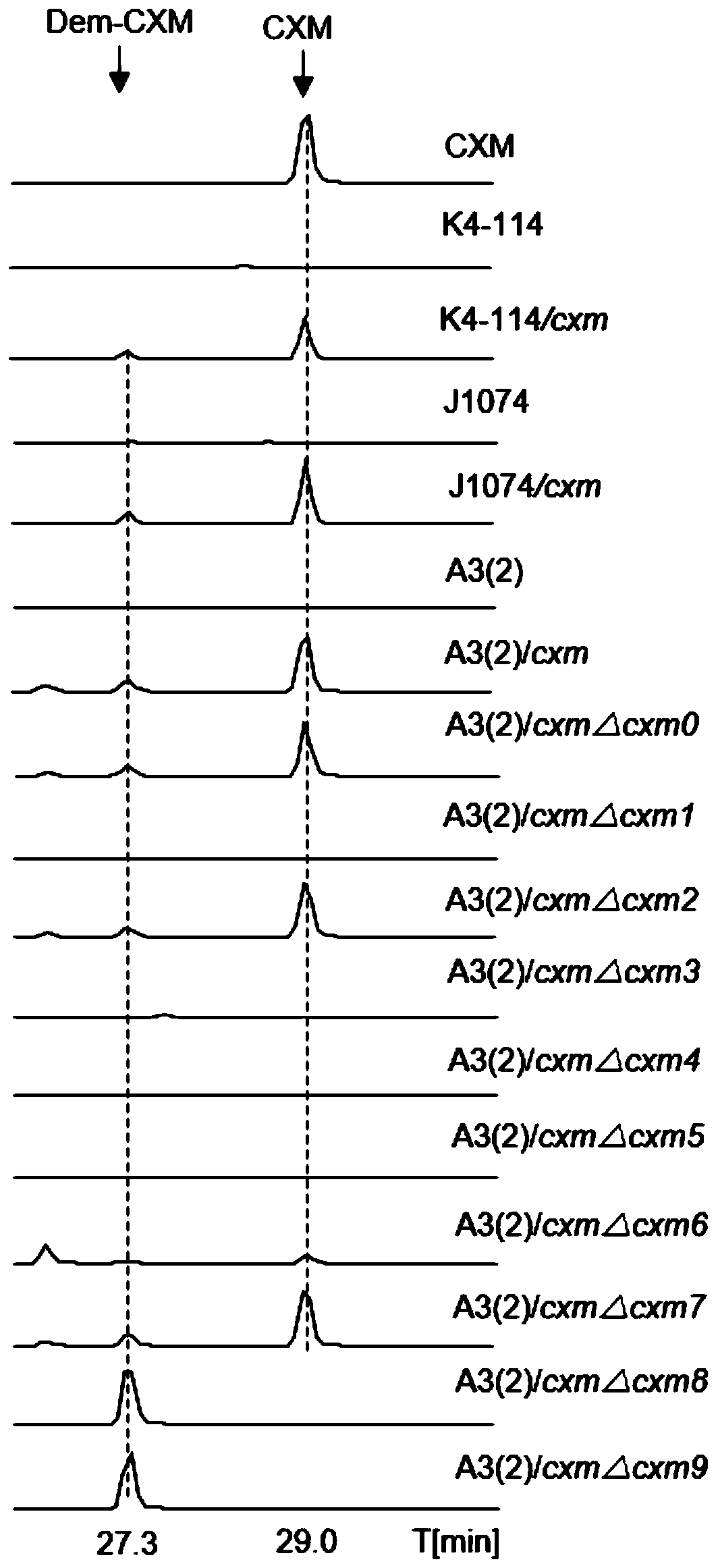
[0058] Using pSET152 as a template, amplify intgrase-attP-oriT-apra carrying a homology arm, and transform the obtained PCR product with the plasmid p15A-cm-cxm directly cloned from the gene cluster into the strain GB05red that has been induced to express the recombinase Line loop recombination, picking clones for enzyme digestion and identification, obtained the combined transfer plasmid p15A-cm-apra-cxm integrated with the noveltymycin gene cluster. apra-cxm moved to Streptomyces coelicolor A3(2) through conjugation transfer and integrated into the attB site of its chromosome, cultured at 37°C for 17h-18h, then mixed with 1 ml of sterile water containing appropriate amount of antibiotics and nalidixic acid (with to inhibit the growth of Escherichia coli) to cover, upside-down culture at 30°C for 7 days to see zygote growth, pick clones for PCR verification, and inoculate the correct clones ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com