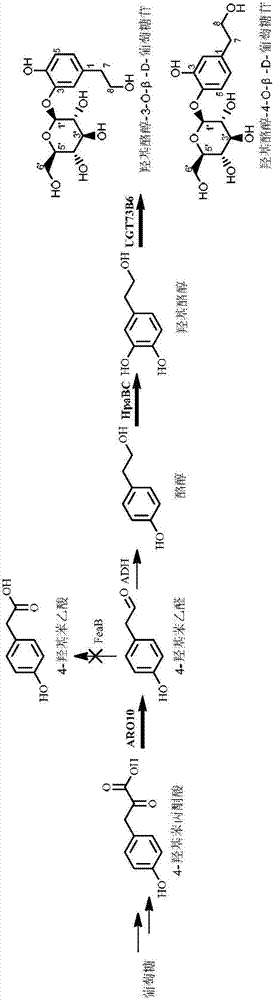
Escherichia coli for expressing hydroxytyrosol and hydroxytyrosol glucoside and construction method and application thereof
A technology of hydroxytyrosol and Escherichia coli, which is applied in the field of bioengineering, can solve the problems of low yield and yield of glycosylated strains, unsafety, complicated process, etc., and achieves easy large-scale fermentation culture, low cost, and rapid growth Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
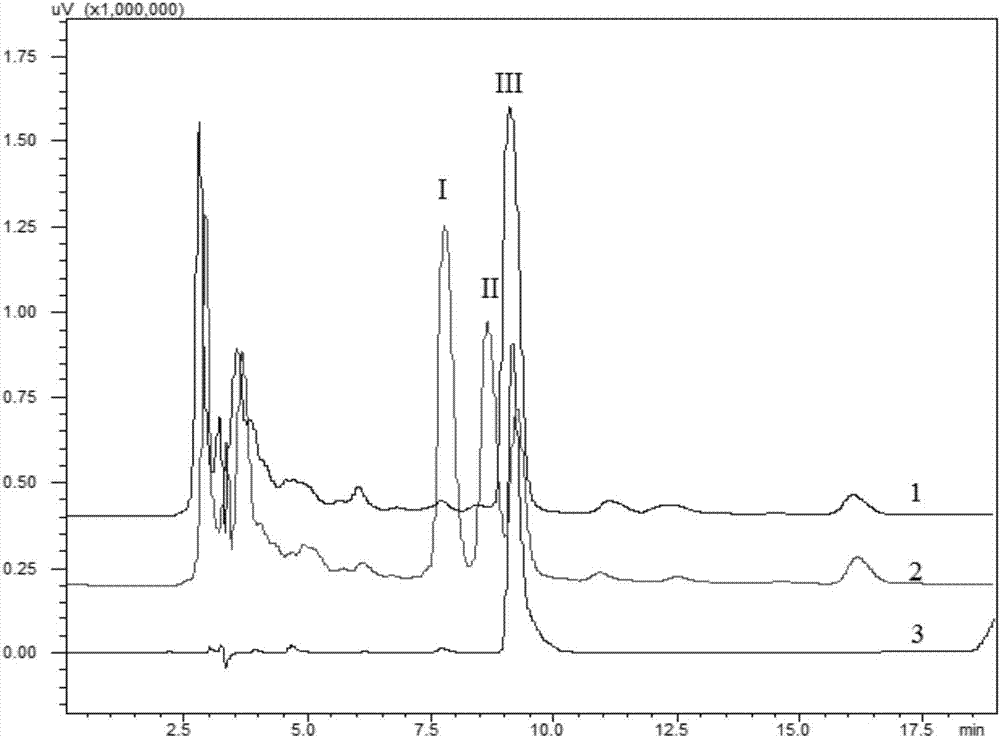
Image
Examples
Embodiment 1
[0036] Gene UGT73B6 or its mutant UGT73B6 FS For the specific acquisition method, please refer to Chinese patent 201510160497.8.
[0037] pET28a-UGT73B6 FS -P Trc ARO10-P Trc HpaBC construction method:
[0038] NotI and XhoI double restriction plasmid pTrcHisB-P Trc ARO10-P Trc HpaBC, obtained with P Trc Promoter Gene Fragment P Trc ARO10-P Trc HpaBC, and NotI and XhoI double-digested vector pET28a-UGT73B6 FS The fragments were ligated and transformed into E.coli DH5α to obtain the intermediate plasmid pET28a-UGT73B6 FS -P Trc ARO10-P Trc HpaBC.
[0039] Among them, pTrcHisB-P Trc ARO10-P Trc The construction method of HpaBC is consistent with that in Chinese patent 201510242626.8 (a method and application of Escherichia coli to produce hydroxytyrosol using a simple carbon source such as glucose); pET28a-UGT73B6 FS The construction method of pET28a-UGT73B6 is consistent with the literature construction method of pET28a-UGT73B6 (Bai Y, Bi H, Zhuang Y, et al. Prod...
Embodiment 2
[0041] Construction method of plasmid pBb0-T7:
[0042] Using Escherichia coli BL21 (DE3) genomic DNA as a template, using primers T7Pol-5FPEcoRI (SEQ ID No: 1) / T7Pol-3RPPstI (SEQ ID No: 2) to amplify the T7 polymerase gene fragment, EcoRI and PstI double enzyme T7 polymerization The enzyme gene fragment and the expression vector pBb0 were transformed into E. coli DH5α to obtain the vector pBb0-T7. Among them, the construction method of pBb0 is consistent with the literature (Bai Y, Bi H, Zhuang Y, et al. Production of salidroside in metabolically engineered Escherichia coli [J]. Scientific reports, 2014, 4: 6640-6647.).
[0043] SEQ ID No: 1 Sequence: CCGGAATTCATGAACACGATTAACATCGCTAAG
[0044] SEQ ID No: 2 Sequence: GTTCTGCAGTTACGCGAACGCGAAGTCCGACTC
[0045] In the above steps, the reaction program of the PCR amplification reaction can be a conventional PCR amplification reaction program, for example, it can be: 94-95°C pre-denaturation for 4-5 minutes; 96-98°C denaturation...
Embodiment 3
[0047] Plasmid pET28a-UGT73B6 FS -P Trc ARO10-P Trc HpaBC-P lacUV5 The construction method of T7:
[0048] Using pBb0-T7 as a template, PCR amplification with P lacUV5 Promoter of the T7 polymerase gene. The PCR primers are T7-5FP-XhoI, whose sequence is shown in SEQ ID No: 3, and T7-3RP-XhoI, whose sequence is shown in SEQ ID No: 4. Use XhoI to single-enzyme digest with P lacUV5 Promoter of T7 polymerase gene and vector pET28a-UGT73B6 FS -P Trc ARO10-P Trc HpaBC, and transformed into E.coli DH5α, the final expression vector pET28a-UGT73B6 was obtained FS -P Trc ARO10-P Trc HpaBC-P lacUV5 T7.
[0049] SEQ ID No: 3 Sequence: CCGCTCGAGTTTACACTTTATGCTTCCG
[0050] SEQ ID No: 4 Sequence: CCGCTCGAGTTACGCGAACGCGAAGTCC
[0051] In the above steps, the reaction procedure of the PCR amplification reaction is the same as in Example 2.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com