Microorganism sample DNA storage solution
A technology of microorganisms and preservation solutions, applied in the field of biological sample preservation solutions, which can solve problems such as poor applicability, unsuitability for clinical testing, and unsuitability for DNA preservation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 Sample source and group storage method
[0023] Due to the high biological preservation requirements of fecal microbial samples, a preferred embodiment of the present invention uses human fecal microbial samples as experimental samples to carry out the following experiments.
[0024] S1. Collect feces samples from 5 volunteers, collect 18 samples from each volunteer, and do 6 different treatments respectively, and repeat the test 3 times for each treatment;
[0025] S2, different processing methods
[0026] a. Freeze for one week (label 0): Collect at least 0.2g of feces samples, quick-freeze in liquid nitrogen, store in a sealed seal at -80°C, store for one week for DNA extraction, 16S rRNA gene V3 / V4 variable region amplification, library construction , sequencing, etc.;
[0027] b. Preservation solution for one week (label 1-3): Collect at least 0.2g of feces samples, use 3 different formulas of the present invention (the ingredients are shown in Table 1)...
Embodiment 2
[0034] Embodiment 2 Fecal sample processing method
[0035] The present embodiment provides a kind of processing method to above-mentioned stool sample:
[0036] S1. Pretreatment of samples in the control group:
[0037] Solid feces (corresponding to sample numbers 0 and 5):
[0038] Weigh 200mg of solid feces into a 2ml centrifuge tube, add 800ul stool DNA Buffer A, fully shake and mix for about 5min, and centrifuge at 1800g for 1min;
[0039] Take out 50ul of the resuspension in a clean 1.5ml centrifuge tube, add 800ul of Lysis-BindingBuffer, vortex and mix, and lyse at 70°C for 5min. After centrifugation for 5 minutes, transfer the supernatant to a clean 1.5ml centrifuge tube.
[0040] S2, experimental group sample pretreatment
[0041] Feces preservation solution (corresponding to sample number 1-4):
[0042] Take 200ul of semi-liquid feces and add no more than 10% (w / v) stool DNA Buffer A to dilute, fully shake for 5min, and take out 50ul of the resuspension into a c...
Embodiment 3
[0058] Example 3 Comparison of the diversity of the sample flora in different preservation methods
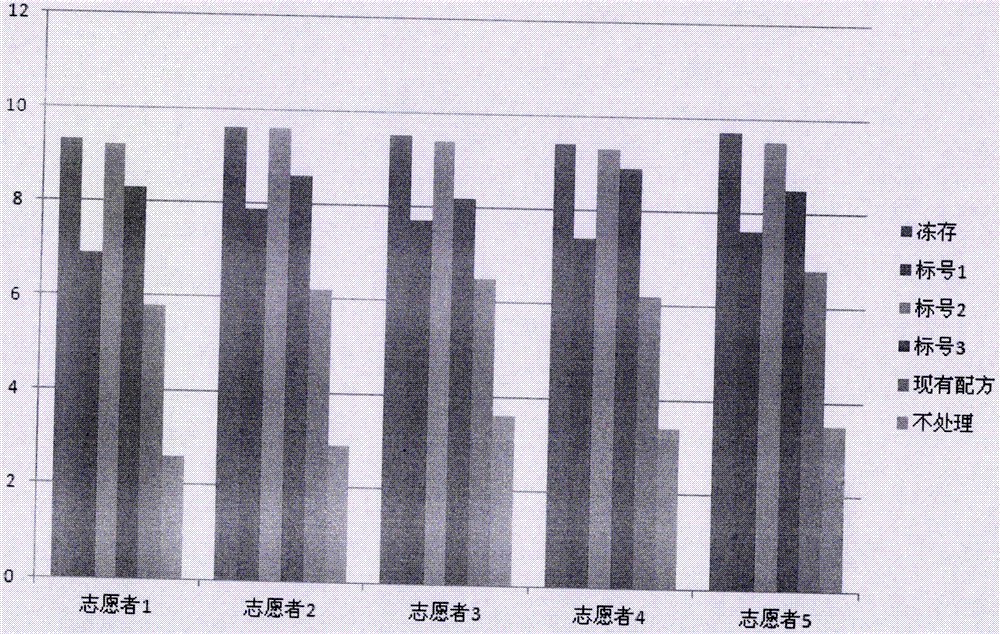
[0059] After the above-mentioned experimental operations, the sequence information of the intestinal flora of 5 volunteers under 6 treatments was obtained, and through bioinformatics analysis, the Shannon diversity index of the flora was calculated and compared. The Shannon index of each volunteer after testing is as follows: figure 1 As shown, the statistical results are shown in Table 3. The higher the value of the Shannon diversity index in the table, the higher the richness of the bacteria, and the p value is the difference between the sample numbered 0-4 and the sample numbered 5. The reliability analysis, the result shows, the sample of label 0-4 can basically satisfy the preservation of feces microbial sample DNA, yet compared with the preservation solution (label 4) of prior art, the treatment effect of microbial sample preservation solution of the present invention Th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| quality score | aaaaa | aaaaa |
| quality score | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com