Quadruple real-time fluorescence PCR method for detecting four types of pathogenic bacteria
A pathogenic bacteria and fluorescence technology, applied in biochemical equipment and methods, microbial determination/inspection, resistance to vector-borne diseases, etc., can solve the problems of complex operation steps, uneven testing personnel ability, etc., and achieve economical operation. time, improving detection accuracy, and the effect of short detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Embodiment 1 self-made sample
[0042] Sample preparation: buy milk powder in a large local supermarket, according to its instructions, take an appropriate amount of milk powder and dissolve it in 100mL of sterilized ultrapure water to make reconstituted milk, and make 3 parts respectively.
[0043] ① Take the first part as the test sample: Inoculate the reconstituted milk with standard strains such as St.aureus (CICC21648), Sa.enterica (CICC 21490), L.monocytogenes (CICC 21633), V.parahemolyticus (CICC21617) and homogenize .
[0044] ②Use the second portion as a negative control: the reconstituted milk was sequentially inoculated with Escherichia coli O157:H7 (Enterohemorrhage E.coli O157:H7, CICC 21530), Shigella Sonnei (CICC 21535 ), Enterobacter sakazakii (Bntorobatesakazakii, CICC 21560), Bacillus cereus (CICC 10876), Campylobacter jejuni (Campylobacter jejuni, CICC 33291) and other standard strains were homogenized.
[0045] ③ Take the third portion as the spike...
Embodiment 4
[0096] Example 4 Enterprise Entrusted Sample --- Detection of Quick-frozen Dumplings
[0097] The list of ingredients includes: wheat flour, water, pork, fungus, mushrooms, onions, soybean protein, soy sauce, dried shrimp, sesame oil, refined vegetable oil, salt, sugar, starch, spices, monosodium glutamate. Grind the dumplings to be tested and take 25g to 225mL of 7.5% sodium chloride broth, buffered peptone water, Listeria broth LB 1 , sodium chloride alkaline peptone water, cultured at 36°C for 18h, 36°C for 8h, 30°C for 24h, and 36°C for 8h to enrich the bacteria. The 4 enrichment solutions were mixed homogeneously.
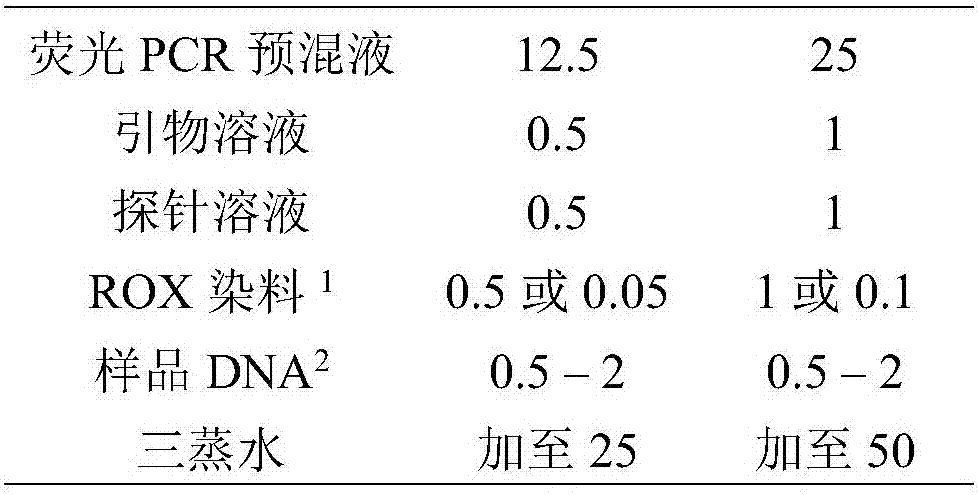
[0098] Using an outsourced DNA extraction kit (TOYOBO -Genome-) to extract DNA from the mixed enrichment solution, the DNA purity is OD 260 / OD 280 =1.81, the DNA concentration was 263.29 ng / μL, and stored at -20°C. Afterwards, the DNA was diluted to 100ng / μL for detection. The primer solution, the probe solution reagent and the positive control were the...
Embodiment 5
[0110] Example 5 Individual entrusted samples --- detection of oysters
[0111] Shred the oyster meat to be tested and take 25g to 225mL of 7.5% sodium chloride broth, buffered peptone water, and Listeria broth LB 1 , sodium chloride alkaline peptone water, cultured at 36°C for 18h, 36°C for 8h, 30°C for 24h, and 36°C for 8h to enrich the bacteria. The 4 enrichment solutions were mixed homogeneously.
[0112] Use an outsourced DNA extraction kit (name: Shanghai Sangong Bacteria Genomic DNA Rapid Extraction Kit) to extract DNA from the mixed enrichment solution, and the DNA purity is OD 260 / OD 280 =1.81, the DNA concentration was 277.27ng / μL, and stored at -20°C. Afterwards, the DNA was diluted to 100ng / μL for detection. The primer solution, probe solution reagent and positive control were the main components of the kit described in this patent. The negative control was plant-derived DNA, and the blank control was triple-distilled water. The fluorescent PCR reagent is Shan...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com