Detection method for identifying American classical PRRSV strain, HP-PRRSV strain and new-type viral NADC30 strain at the same time
A technology of classic strains and mutant strains, applied in the field of detection and molecular biology detection, can solve problems such as low accuracy, PCR product contamination, inability to distinguish PRRSV, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072]1. Design of specific primers and specific probes: Download multiple American PRRSVs from Genbank (classic strains HN1, SD9521, VR2332, SD1100, CH1a; highly pathogenic variant strains SX1, TJ, JXA1, HUN4; NADC30like : HNyc15, HNjz15, JL580, HENXX1 and SDlz1601) NSP2 gene and N gene sequence, use BioEdit software package to carry out Clustalw comparison, utilize Primer 3.0 software to design specific TaqMan probe, primer according to different virus types. According to the conserved region of the N gene sequence of American-type and European-type PRRSV, the universal primers and universal probes of American-type PRRSV were designed.
[0073] 2. Synthesis of primers and probes: Synthesized by Shanghai Jierui Biological Co., Ltd.
[0074] 3. See Table 2 for primers, probe sequences and probe modifications.
[0075] Table 2 Primers and sequence information
[0076]
[0077]
[0078] 4. Sample template preparation
Embodiment 2
[0084] Example 2 Verification of multiple real-time fluorescence methods
[0085] 1. Specificity verification
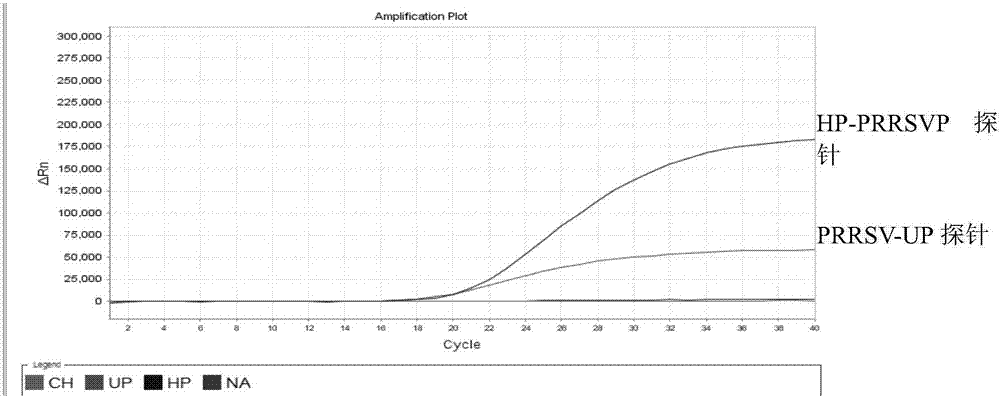
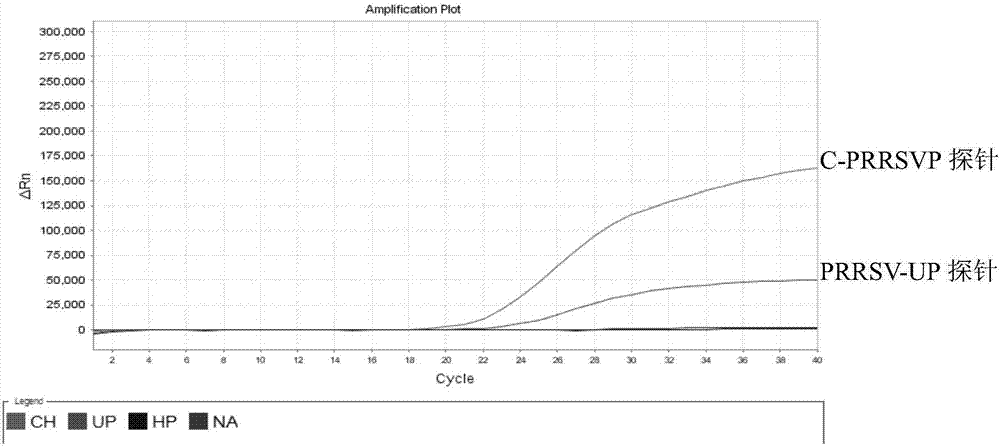
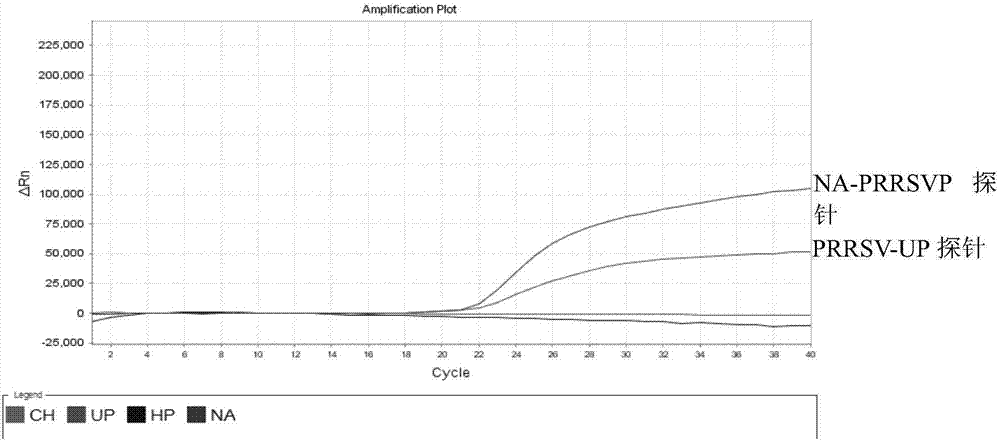
[0086] Utilize the kit method of the present invention to respectively use NADC30like LZ, highly pathogenic SX-1, classic SD-1 strain, vaccine virus R98 strain, TJM-F92 strain, JXA1-R, and swine fever virus, porcine parvovirus, porcine Epidemic diarrhea virus, porcine pseudorabies virus and porcine circovirus type 2 virus were used as templates for multiplex real-time fluorescent PCR amplification to verify the specificity of their primers and probes. The results are shown in Table 3, and the results show that the designed primers and probes of the present invention have strong specificity.
[0087] Table 3. Specificity Verification Tests
[0088]
[0089]
[0090] 2. Sensitivity evaluation
[0091] Quantify NADC30like, highly pathogenic, classic strain positive standards to 10 7 copy / μL, serially diluted 10 times to 1.0×10 6 , 1.0×10 5 , 1.0×10 4 , 1.0×...
Embodiment 3
[0092] Example 3 Clinical Suspect Sample Detection
[0093] The multiple real-time fluorescent RT-PCR detection method established by the present invention for the classic type, highly pathogenic variant strain and NADC30like strain of PRRSV simultaneously detects 30 clinically suspicious samples, and the sample types include pig lung, lymph node, tissue and serum. Detection was performed using both virus isolation methods and sequencing. The results are shown in Table 4, and the results show that the method established by the present invention is completely consistent with the verification of the sequencing results after virus isolation, and the method is accurate and reliable.
[0094] Table 4 Test results of clinical samples
[0095]
[0096]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com