Application of soybean transcription factor GmWRKY23 gene in stress resistance
A transcription factor and transgenic technology, applied in the field of genetic engineering and botany, to achieve obvious antibacterial effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
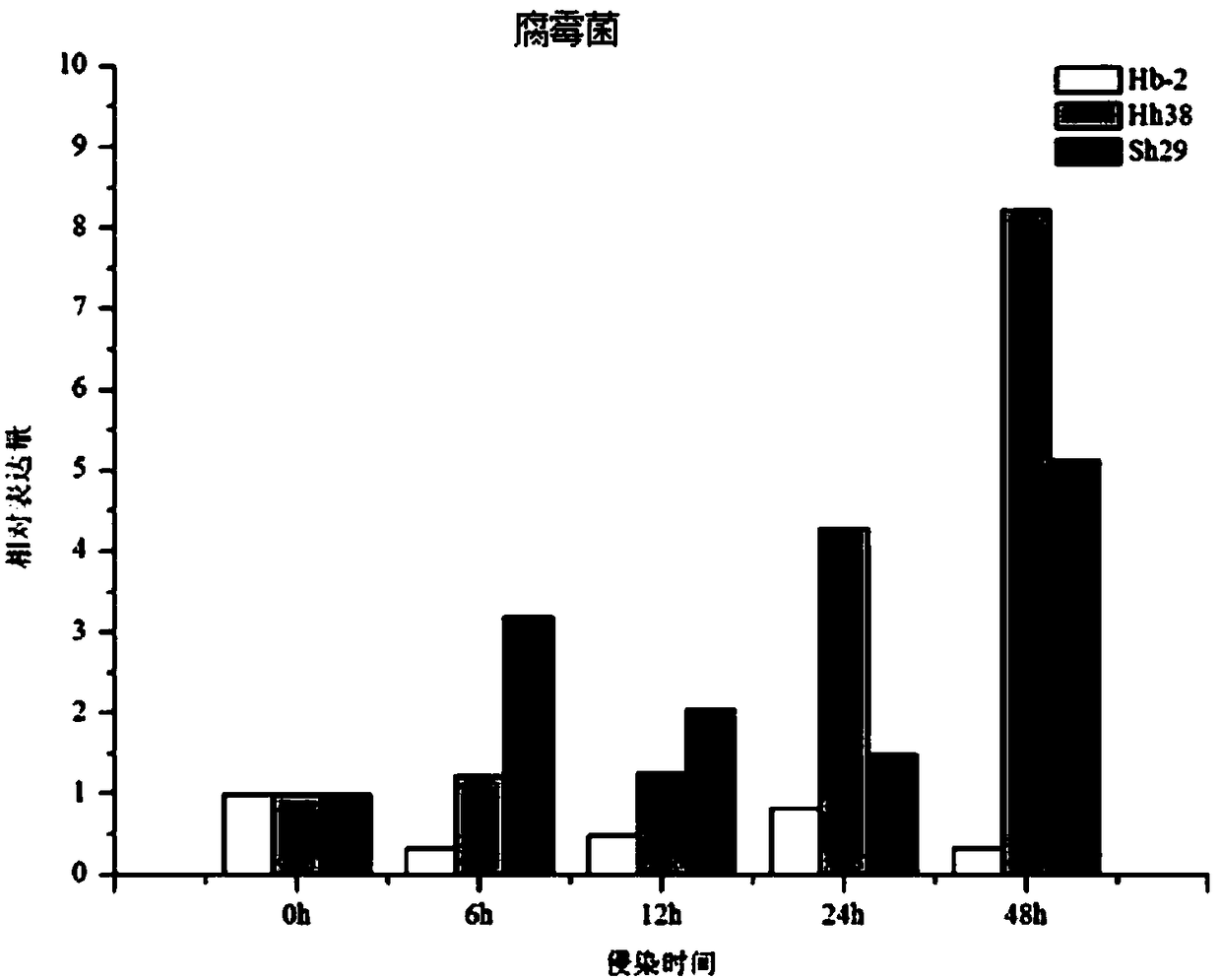
[0051] Example 1 Expression pattern of GmWRKY23 in young roots of different resistant cultivars under Pythium infection
[0052] Soybean germplasm Huibuzhidou (Hb-2), Heihe 38 (Hh38), and Suinong 29 (sh29) were provided by the Soybean Research Institute of the Heilongjiang Academy of Agricultural Sciences; Pythium was provided by the Soybean Research Institute of the Heilongjiang Academy of Agricultural Sciences.
[0053] Ⅰ. Soybean germination test
[0054] Wash the soybean seeds of Hb-2, Hh38 and sh29 with distilled water, dry them, and sterilize them with chlorine gas for 12-16 hours; then soak them in sterile deionized water for 12 hours, and place them evenly in a container with WA medium (water agar medium) on the plate, about 2cm away from the edge of the plate, placed in a growth chamber at room temperature for germination culture.
[0055] Ⅱ. Culture of Bacteria
[0056] Using a sterilized hole puncher, from the V8 medium for cultivating Pythium (V-8 fruit juice 200...
Embodiment 2
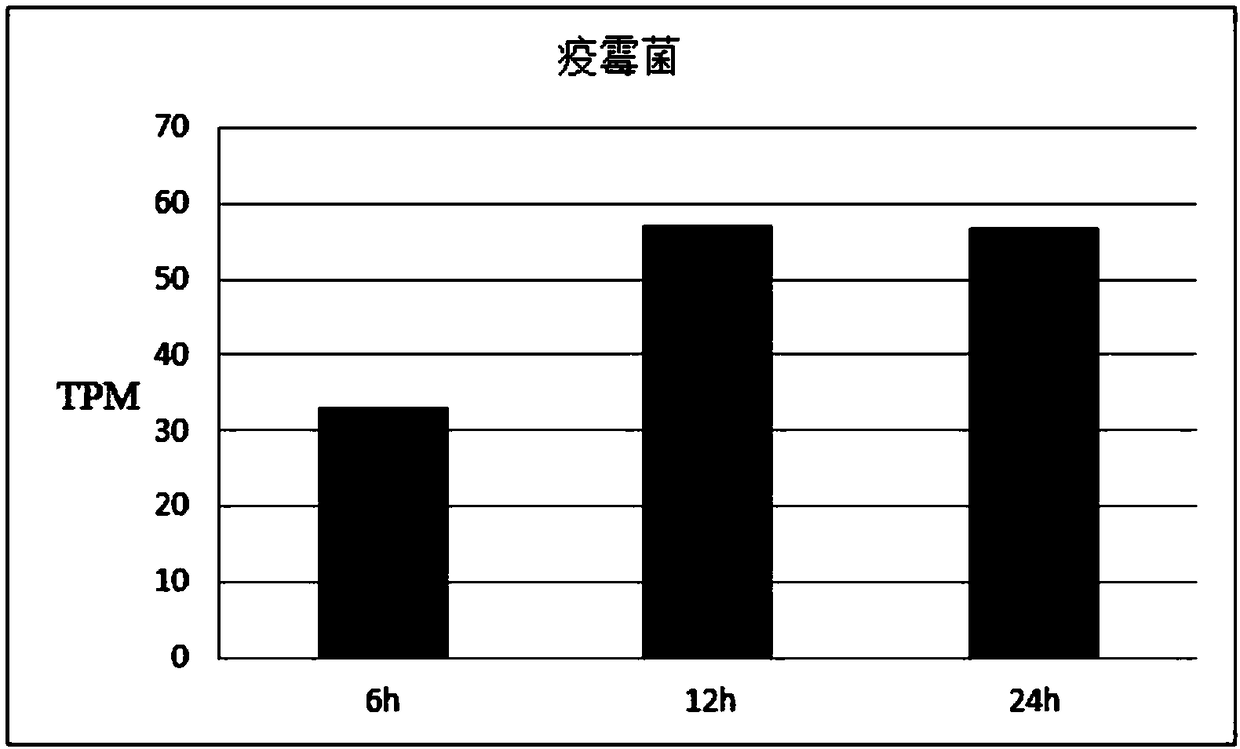
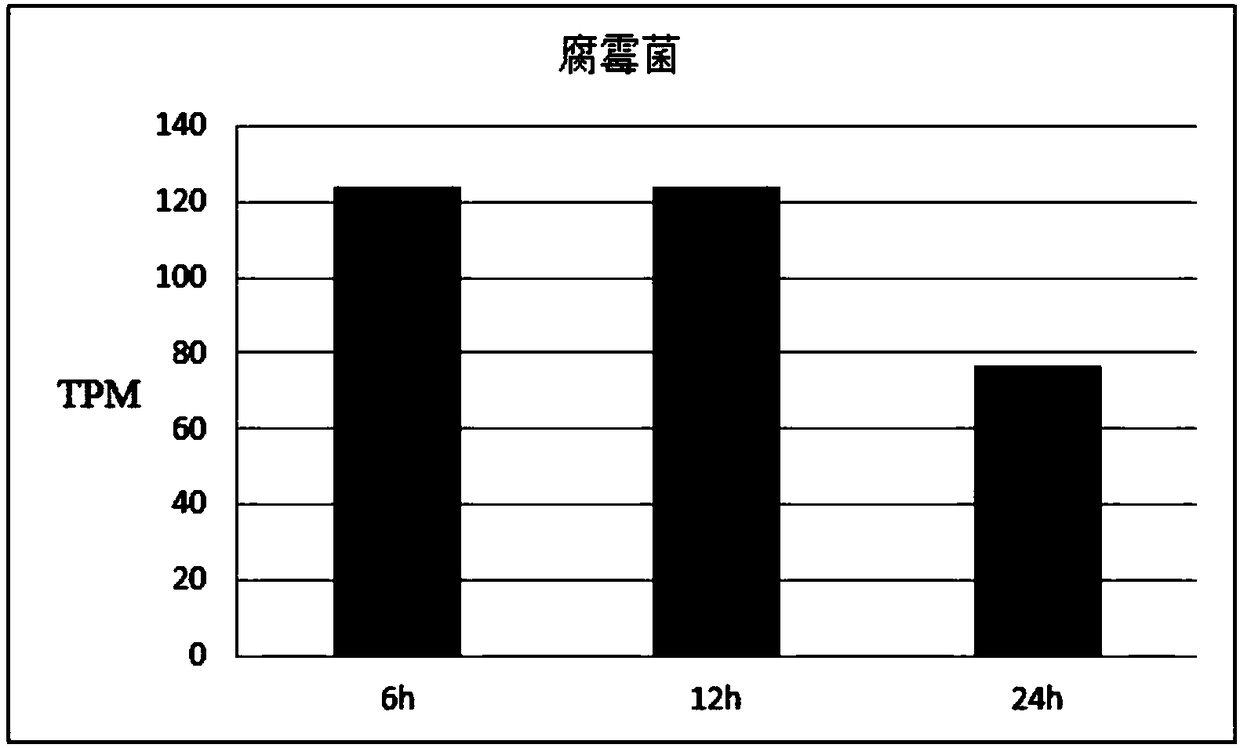
[0064] Example 2 The expression pattern of GmWRKY23 gene under the infection of different root rot pathogens
[0065] Rhizoctonia solani was provided by the Soybean Research Institute of Heilongjiang Academy of Agricultural Sciences; Phytophthora was provided by the Plant Protection Office of Hejiang Branch of Heilongjiang Academy of Agricultural Sciences.
[0066] The disease-resistant variety Hb-2 was used as a test material to explore the gene expression profiles of Phytophthora, Pythium, and Rhizoctonia solani infecting young soybean roots. The specific test methods refer to Example 1. The result is as Figure 2-4 , where TPM (Transcript per million clean Tags) is the number of copies of the transcript per million clean Tags. The expression level of GmWRKY23 gene was the highest after 6 hours of Pythium infection, and the expression level decreased at 24 hours; under the infection of Phytophthora, the expression level of GmWRKY23 in soybean young roots was the highest at ...
Embodiment 3
[0067] Cloning of embodiment 3 GmWRKY23 gene
[0068] Ⅰ. Extraction of RNA
[0069] The RNAprep pure plant total RNA extraction kit from TIANGEN Company was used to extract the total RNA of the hypocotyls of the soybean variety Hb-2 inoculated with Pythium, and the purity of the extracted total RNA was tested.
[0070] (1) The tweezers, medicine spoon and mortar used in the test were first grilled with alcohol for 15 minutes. The tips and centrifuge tubes for RNA extraction are RNase-Free products from Axygen Company.
[0071] (2) Weigh 100 mg of the hypocotyl tissue of soybean variety Hb-2 6 hours after inoculation with Pythium, quickly grind it into powder in liquid nitrogen, add 500 μL SL (check whether β-mercaptoethanol has been added before use), and immediately vortex Shake vigorously to mix; centrifuge at 12,000rpm (~13,400×g) for 2min.
[0072] (3) Transfer the supernatant to the filter column CS (the filter column CS is placed in the collection tube), centrifuge at...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com