A kind of endogenous l-asparaginase II gene knockout host bacteria, its preparation method and application
A technology of asparaginase and exogenous genes, applied in the field of preparation of Escherichia coli host strains, can solve the problems of ansB gene and host bacterial genome instability, low recombination efficiency, and treatment failure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Example 1: Knockout of the ansB gene on the BL21 (DE3) genome
[0055] 1. Primer Design
[0056] According to the upstream and downstream sequences of the ansB gene on the BL21 (DE3) genome, primers for the upstream and downstream homologous recombination arms, connecting primers and identification primers for the upstream and downstream homologous recombination arms were designed. The primer sequences are shown in Table 3.
[0057] Table 3 Primer Sequence
[0058]
[0059]
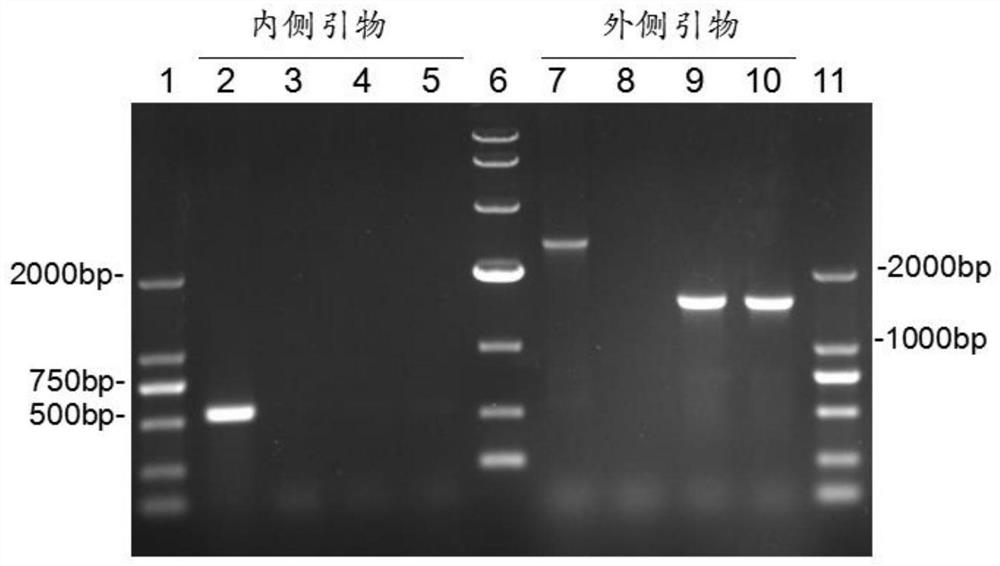
[0060] 2. Amplification of targeting sequences and construction of targeting vectors
[0061] Using a genome extraction kit ( Genomic DNA Purification Kit, Promega) extracts the genome of BL21 (DE3) as a PCR template, primers are upstream and downstream homologous recombination arm primers (SEQ ID NO: 7 to SEQ ID NO: 10), using high-fidelity DNA polymerase (PlatinumTaq , Invitrogen) to amplify the upstream and downstream homologous recombination arms. Using the connecting primer (SEQ ID N...
Embodiment 2
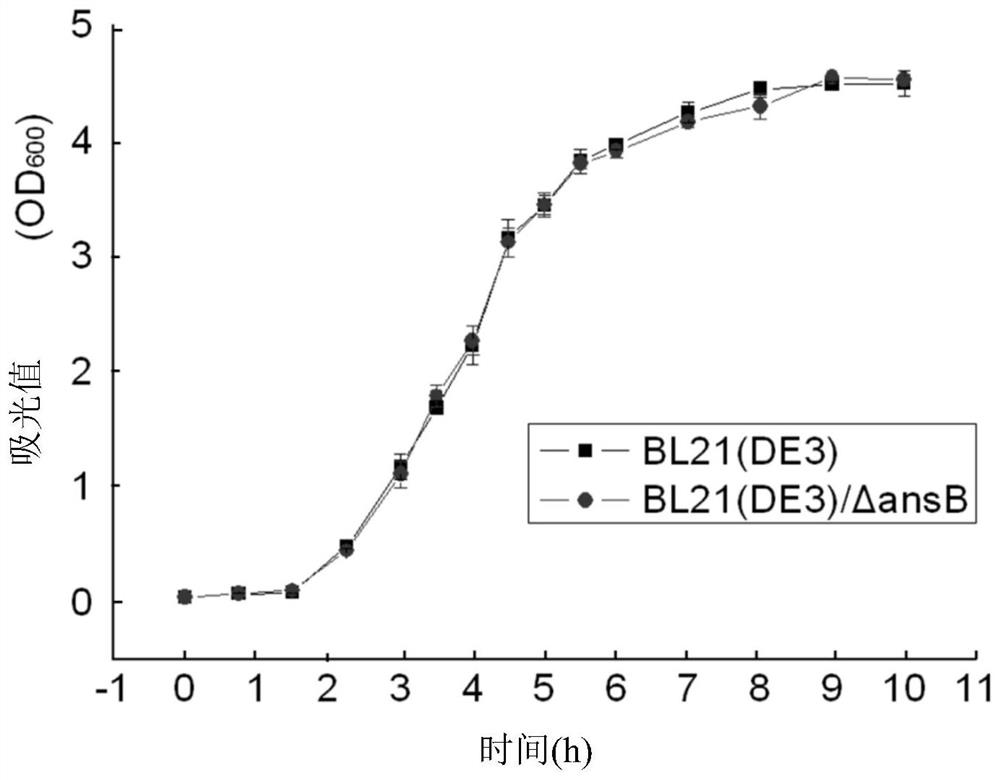
[0068] Embodiment 2: Comparison of the growth characteristics of wild bacteria BL21 (DE3) and gene knockout bacteria BL21 (DE3) / ΔansB
[0069] Prepare LB medium, the formula is 1% Tryptone, 0.5% yeast extract, 1% NaCl. Inoculate in 5mL seed medium with 0.5% ratio, 50mL micro bioreactor (Mini Bioreactor, Corning), 37°C, 220rpm culture to logarithmic growth phase, transfer to 50mL LB medium with 2% ratio, do 3 parallel A 250mL shake flask (ErlenmeyerFlask, Corning), sampling every 30min or 1h, using a UV spectrophotometer to detect OD 600 value. Taking time as the abscissa, OD 600 Values plot growth curves on the ordinate, see figure 2 .
[0070] Formula 1:
[0071]
[0072] In the formula, μ is the specific growth rate in h -1 , T2-T1 is the time spent by microorganisms from time point T1 to time point T2, in h, N1 is the cell mass of microorganisms at time point T1, and N2 is the cell mass of microorganisms at time point T2.
[0073] The obtained data was processe...
Embodiment 3
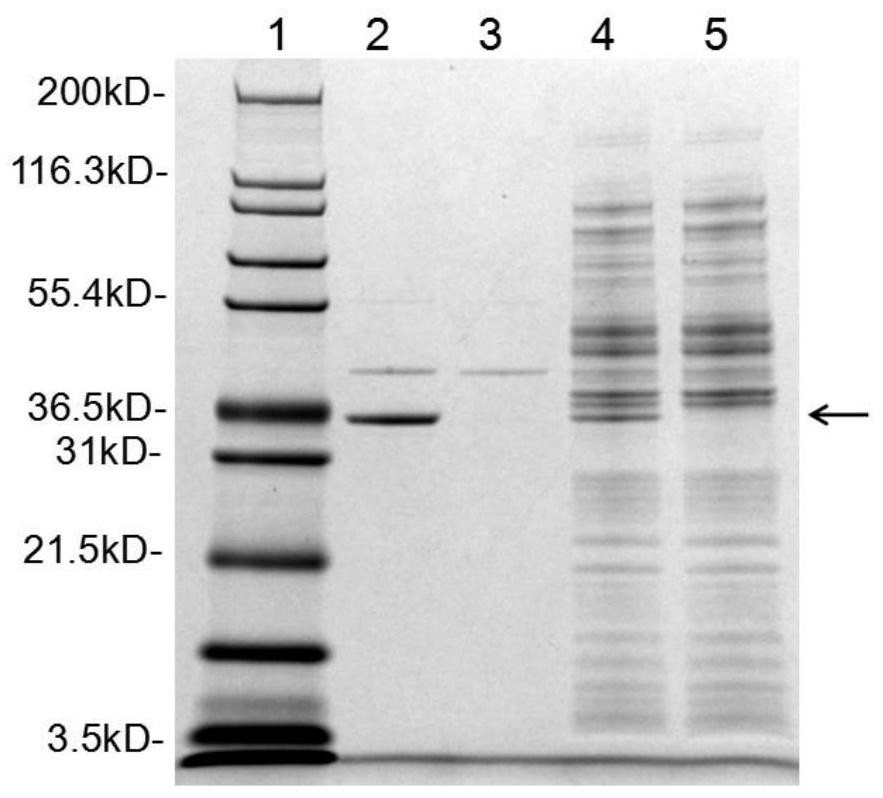
[0074] Example 3: Background expression of L-asparaginase II of wild-type bacteria BL21 (DE3) and gene knockout bacteria BL21 (DE3) / ΔansB
[0075] Prepare seed medium with the formula of 1% Tryptone, 0.5% yeast extract, 1% NaCl, 0.1% L-asparagine, prepare background expression medium with the formula of 1% Tryptone, 0.5% yeast extract, 1% NaCl, 0.6% L-asparagine. Get glycerol bacteria, inoculate 5mL seed culture medium with 0.1% ratio, 50mL miniature bioreactor (Mini Bioreactor, Corning), 25 ℃, 150rpm cultivate to OD 600 = around 4 (16-18 hours). Transfer to 30 mL background expression medium at a ratio of 4%, culture in a 125 mL flat-bottomed shaker flask (Erlenmeyer Flask, Corning) at 25° C., 150 rpm for 14 hours. Centrifuge at 8000rpm for 5min to collect the bacterial cells, extract periplasmic protein by osmotic pressure method (refer to pET manual), use 10kD concentration tube (Spin- UF 20, Corning) were properly concentrated and analyzed by SDS-PAGE electrophoresis. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com