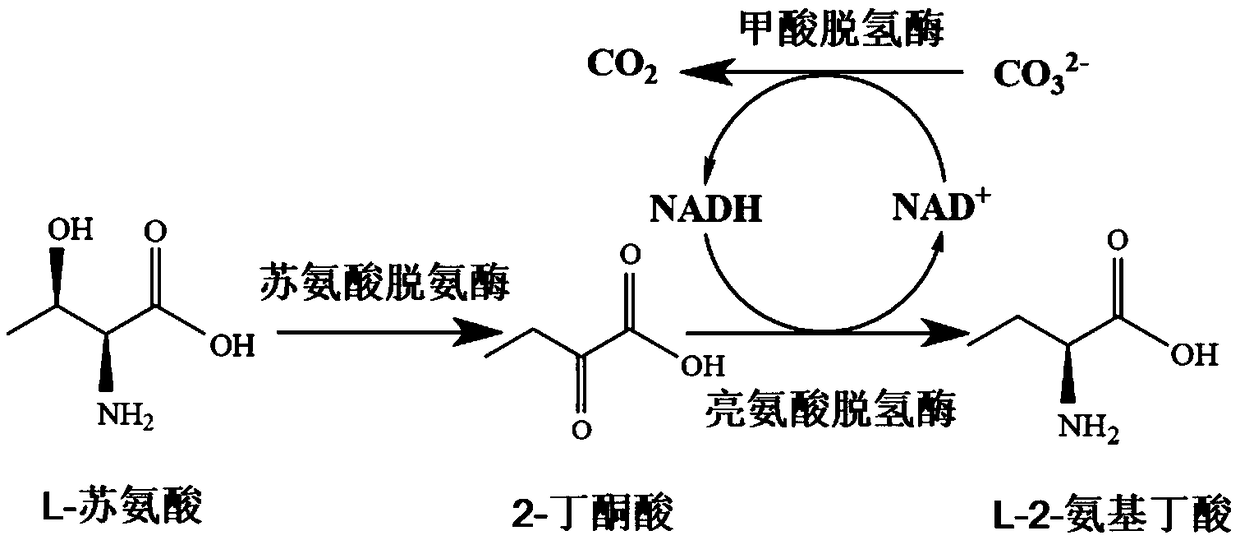
Construction and application of recombinant strain converting L-threonine to L-2-aminobutyric acid
A technology of aminobutyric acid and threonine deaminase, applied in the direction of microorganism-based methods, bacteria, microorganisms, etc., can solve the problems of low efficiency of coenzyme cycle regeneration, complex multi-enzyme addition, long conversion time, etc., and achieve low cost , high product optical purity and high conversion efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1: Acquisition of genetically engineered bacteria producing Escherichia coli threonine deaminase
[0034] (1) Escherichia coli W3110 was inoculated in LB medium, cultured at 37°C for 12 hours to collect bacteria, and genomic DNA was extracted using a bacterial genome extraction kit.
[0035](2) Using primers EcTD-1 (5'CGGGATCCATGGCTGACTCGCAACCCCTG 3', SEQ ID NO:8) and EcTD-2 (5'CCCAAGCTTCTAACCCGCCAAAAAGAACCTGAAC 3', SEQ ID NO:9) to clone threonine deaminase from the genome Gene EcTD;
[0036] (3) Connect the target gene to the PMD19simple cloning vector for sequencing, select the correct gene fragment and digest it with BamHI and XhoI, and then connect it to the plasmid pET28a that has been double digested with the same two enzymes;
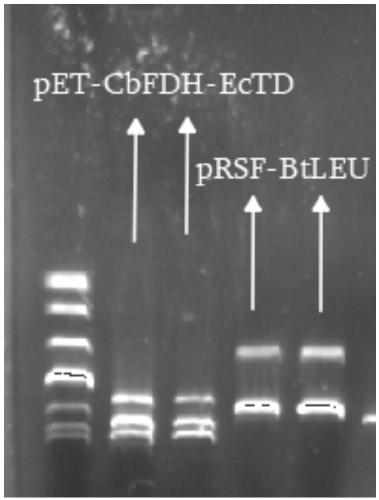
[0037] (4) Introduce the constructed expression plasmid into E.coli BL21(DE3), screen and verify it on the LB plate containing kanamycin, and select the strain with the correct target gene. The gene sequence is SEQ ID NO:1 , the ...
Embodiment 2
[0040] Example 2: Acquisition of genetically engineered bacteria producing Bacillus thuringiensis leucine dehydrogenase
[0041] (1) Bacillus thuringiensis was inoculated in LB medium, cultured at 37°C for 12 hours to collect the bacteria, and the genomic DNA was extracted using a bacterial genome extraction kit.
[0042] (2) Bacillus thuringiensis leucine was cloned from genomic DNA using primers BtLeuDH-1 (5'CGGGATCCATGCGCGTTATGGTCTTG 3', SEQ ID NO:10) and BtLeuDH-2 (5'CCCAAGCTTTTAGCGACGGCTAATAATATCGTG 3', SEQ ID NO:11) respectively Acid dehydrogenase gene BtLeuDH.
[0043] (3) Connect the target gene to the PMD19simple cloning vector for sequencing, select the correct gene fragment to digest with BamHI and XhoI, and connect it to pET28a that has been double-digested with the same two enzymes;
[0044] (4) Introduce the constructed expression plasmid into E.coli BL21(DE3), screen and verify it on the LB plate containing kanamycin, and select the strain with completely corre...
Embodiment 3
[0047] Embodiment 3: the acquisition of the genetic engineering bacterium that produces formate dehydrogenase
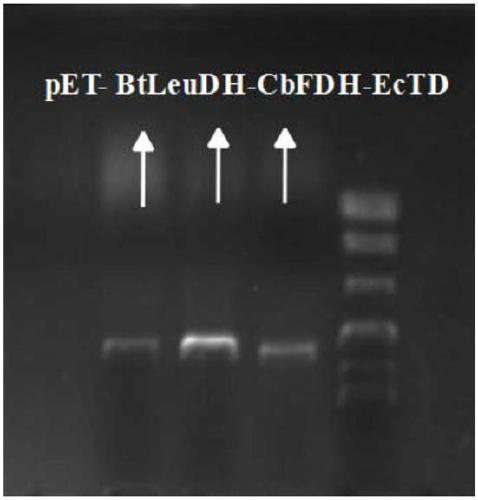
[0048] The codon optimization of the formate dehydrogenase gene (FDH) derived from Candida boidinii (original gene sequence such as SEQ ID NO: 3) makes the gene sequence more suitable for the large intestine expression system. After optimization, the gene sequence is such as SEQ ID NO: 4, and the amino acid sequence is as follows: SEQ ID NO:7. The artificially synthesized FDH gene fragment containing BamHI and XhoI restriction sites was double-digested, and then connected with the expression vector pET28a obtained by the same double-digestion to construct the recombinant plasmid pET28a-FDH, and the recombinant plasmid was transformed into the expression host E. In coli BL21(DE3), screening and verification were carried out on the LB plate containing kanamycin, and the correct positive strain was screened out.
[0049] Inoculate the above codon-optimized engineered b...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com