An efficient and economical method for extracting DNA from soil microorganisms
A soil microorganism and extraction method technology, which is applied in the field of rapid extraction of soil microbial DNA, can solve the problems of poor effect of the method and expensive foreign kits, and achieve the goal of saving reagent consumption, simplifying the DNA collection process, and reducing the amount of use Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
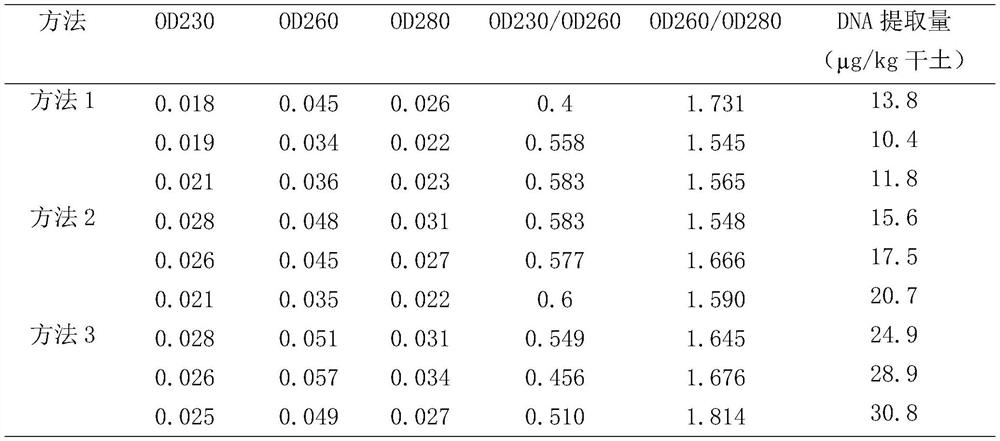
[0027] Embodiment 1 (three repetitions)
[0028] The soil extraction kit Soil DNA Isolation Kit of mobio company was used to extract, and the method was as follows:
[0029] Add 2g soil to 15mL Bead Tube, add 0.25mLSR1 and 0.8mLSR2, add 2.5mL BeadSolution to Bead Tube, then add 3.5mL phenol: chloroform: isoamyl alcohol = 25:24:1 to the test tube, cover and vortex Blend until the layering disappears. Vortex at the maximum speed for 15 minutes, centrifuge at 2500g for 10 minutes at room temperature. After removal, carefully transfer the upper aqueous phase to a clean 15mL collection tube and discard the lower phenol. Add 1.5mL SR3 to the water phase, then vortex and mix well, incubate at 4°C for 10min, room temperature, centrifuge at 2500g for 10min. Transfer the supernatant to a new 15mL collection tube, add 5mL SR4 solution to the collection tube containing the supernatant, mix well and incubate at room temperature for 30min. Centrifuge at 2500g for 30min at room temperatu...
Embodiment 2
[0030] Embodiment 2 (cold-thaw method currently used)
[0031]Take 1g of the ground soil powder, put it in a 10mL centrifuge tube, add 2mL of sodium phosphate buffer (0.12mol / L, pH8.0), mix it, put it on a shaker at 30°C, and shake it at 150r / min for 15min. 8 000r / min, centrifuge for 10min. Take the precipitate and repeat the above operation. Take the precipitate, add 1.5mL of Lysis Solution I (0.15mol / LNaCl, 0.1mol / LEDTA, pH8.0) and 0.5mL of 50mg / L lysozyme, mix well, bathe in 37℃ water for 2h, and shake every 20-30min. Add 2 mL of lysate II (0.1 mol / L NaCl, 0.5 mol / L Tris-HCl, pH 8.0, 10% SDS), freeze and thaw three times, and centrifuge at 8 000 r / min for 15 min. The supernatant was mixed with an equal volume of phenol reagent (v (phenol): v (chloroform): v (isoamyl alcohol) = 25:24:1), centrifuged at 8 000 r / min for 10 min. Repeat the previous step. Take the aqueous phase and mix it with an equal volume of chloroform and isoamyl alcohol mixture (v(chloroform:v(isoamyl ...
Embodiment 3
[0033] Weigh 1 gram of soil, add 2ml of suspension buffer (sodium phosphate aqueous solution, 0.15mol / L, pH8.0), put 1 gram of glass beads with a particle size of 0.1mm into the extraction tube, and crack the soil under the friction of the glass beads All organisms in the medium include free DNA of G+ bacteria, yeasts, fungi, algae, nematodes, and even eubacterial spores, spores, animal and plant remains, etc.; add humic acid adsorbent 500 μl: it consists of nano-TiO 2 : 0.2M; activated carbon: 0.1% (mass percentage). The specific configuration process is: Weigh 3.2000g spectroscopically pure titanium dioxide into a container, add 12.8g ammonium sulfate, 32mL 98% sulfuric acid and weigh 0.2g activated carbon, heat to dissolve, cool to room temperature, add water to dilute to 200mL, shake well, and set aside . ) mixed fully and shaken for 1min; add 10μL of proteinase K (10mg / mL) solution, shake for 10min after mixing, then add 2ml of high-efficiency lysate (2% CTAB (mass perce...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com