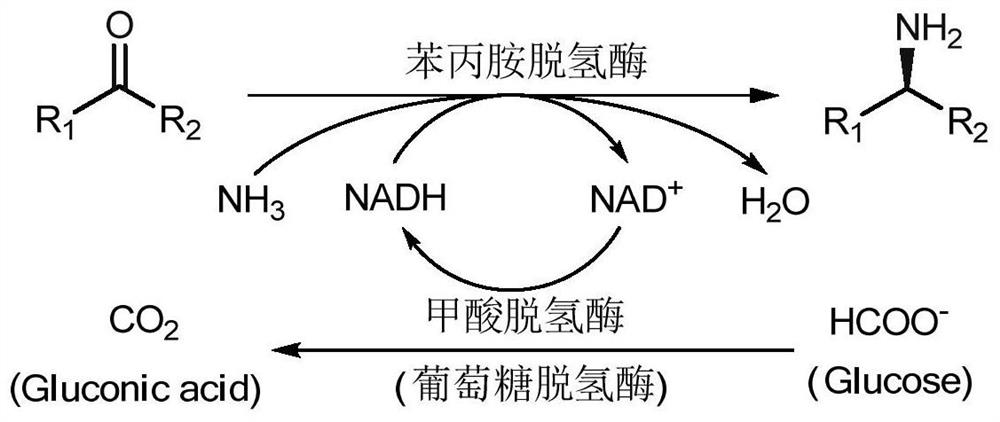
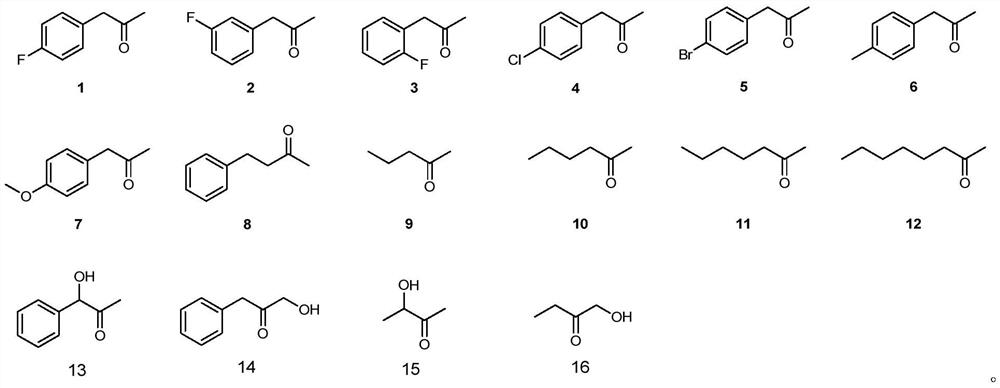
A mutant of amine dehydrogenase and its application in the synthesis of chiral amines and aminoalcohols
A technology of amine dehydrogenase and mutants, applied in the field of amine dehydrogenase mutants and its application in the synthesis of chiral amines and amino alcohols, can solve the problem of low activity of amine dehydrogenase, and achieve wide application value Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] The gene cloning of embodiment 1 phenylalanine dehydrogenase GkPheDH
[0049] According to the open reading frame of phenylalanine dehydrogenase GkPheDH, design upstream and downstream primers:
[0050] The upstream primer, as shown in SEQ ID No.4, comprises a restriction endonuclease BamH I enzyme cutting site;
[0051] The downstream primer, as shown in SEQ ID No.5, contains a restriction endonuclease Hind III enzyme cutting site.
[0052] PCR amplification was performed using the genomic DNA of Geobacillus kaustophilus as a template. PCR system: 2×Taq PCR MasterMix 25 μl, upstream primer and downstream primer (10 ng / μl) 1.5 μl each, genomic DNA (100ng / μl) 1 μl and ddH 2 O 21 μl. The PCR amplification program was: pre-denaturation at 95°C for 5 min, followed by 32 cycles of denaturation at 94°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 60 s, and finally extension at 72°C for 10 min. After the PCR amplification product was purified by gel electrop...
Embodiment 2
[0053] Example 2 Preparation of Phenylalanine Dehydrogenase Recombinant Expression Plasmid and Recombinant Expression Transformant
[0054] The phenylalanine dehydrogenase target gene fragment obtained by PCR amplification in Example 1 and the pET 28a plasmid were simultaneously digested overnight with restriction endonucleases BamH I and Hind III, and then subjected to agarose gel electrophoresis Purification and DNA kit recovery. Under the action of T4 DNA ligase, ligate the recovered target gene and the restriction fragment plasmid of the plasmid at 16°C for 24 hours to obtain the recombinant expression plasmid pET28a-GkpheDH of phenylalanine dehydrogenase, and then transform the recombinant expression plasmid into Cultivate E.coli BL21(DE3) and pick positive clones to obtain recombinant expression transformant E.coliBL21(DE3) / pET28a-GkpheDH.
Embodiment 3
[0055] Example 3 Amine dehydrogenase mutant GkAmDH K78S / N276L Creation and expression of recombinant proteins
[0056] Using the recombinant expression plasmid pET28a-GkpheDH constructed in Example 2 as a DNA template, the 78th amino acid mutation primer was used to mutate the 78th lysine in the amino acid sequence to serine by whole plasmid PCR. The obtained PCR product was degraded by restriction endonuclease Dpn I to remove the DNA template, and after purification, it was used as a template for amino acid mutation at position 276. Using the primers for amino acid mutation at position 276, the asparagine at position 276 of the amino acid sequence was converted to asparagine by whole plasmid PCR. Mutate to leucine, and then degrade the DNA template by Dpn I enzyme, transform the mutant plasmid into E.coli BL21(DE3), you can get the 78th lysine mutation to serine, and the 276th asparagine mutation to Leucine amine dehydrogenase, named GkAmDH K78S / N276L .
[0057] The upstre...
PUM
| Property | Measurement | Unit |
|---|---|---|
| purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com