Marine streptomyces niveus chitosanase gene and application thereof
A technology of chitosanase and streptomyces, applied in application, glycosylase, genetic engineering and other directions, can solve the problems of uncontrollable range of polymerization degree and poor specificity of products
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] The construction of embodiment 1 chitosan hydrolase SnCSN expression bacterial strain
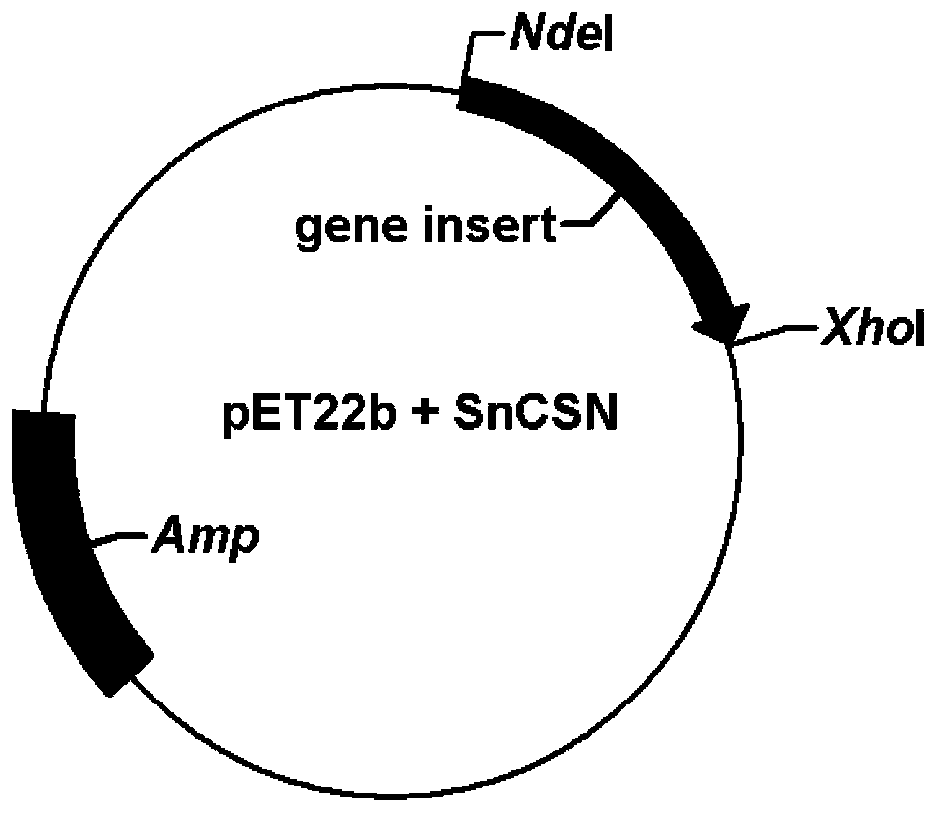
[0035] With reference to the gene encoding chitosan hydrolase SnCSN (GenBank No: AQU65829.1), the present invention entrusts Huada Gene Co., Ltd. to synthesize the gene encoding chitosan hydrolase (excluding signal peptide) through codon optimization. 765 bases, the nucleotide sequence is shown in SEQ ID NO.1, the cloning vector is pET22b, the cloning sites BamHI and XhoI, the carrier resistance is ampicillin (Amp), and the optimized species E.coli ( figure 1 ). Escherichia coli DH5α carrying the expression plasmid pET22b-SnCSN was cultured, the plasmid was extracted using the Plasmid Mini Kit I kit, and then the expression plasmid was introduced into competent Escherichia coli BL21(DE3) to obtain a recombinant strain. The amino acid sequence is shown as SEQ ID NO.2, which contains 255 amino acids, and the predicted protein molecular weight is 29.8kDa.
[0036] SEQ ID NO.1:
[003...
Embodiment 2
[0057] Expression and detection of embodiment 2 chitosan hydrolase SnCSN
[0058] (1) Prepare LB medium containing Amp resistance: 5g / L yeast extract, 10g / L tryptone, 10g / L sodium chloride, sterilize at 120°C for 20min, cool to room temperature and add Amp to make the final concentration 100μg / L mL.
[0059] (2) Inoculate the recombinant strain obtained in Example 1 onto solid LB medium containing Amp resistance, culture overnight at 37°C, pick a single colony and inoculate it into 10 mL of liquid LB culture containing Amp resistance, at 37°C, After culturing on a shaker at 200rmp for 24 hours, inoculate the above-mentioned bacterial solution into 600ml liquid LB culture containing Amp resistance, and cultivate to OD at 37°C and 200rmp bed 600nm When =0.6, add 0.5mM IPTG, induce expression at 16°C, 200rmp for 12 hours, centrifuge at 4000rmp, and collect the bacteria.
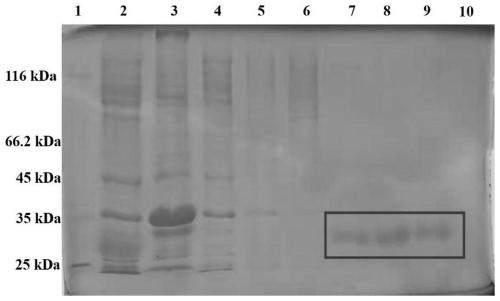
[0060] (3) A small amount of bacterial cells were taken for SDS-PAGE analysis, and the expression of the ta...
Embodiment 3
[0061] Expression, purification and detection of embodiment 3 chitosan hydrolase SnCSN
[0062] (1) Example 2 was collected into thalli, suspended in buffer solution A (50mM Tri-HCl, pH 7.9, 500mMNaCl) and carried out sonication, 12,000rmp was centrifuged to collect the supernatant, and carried out SDS-PAGE detection ( figure 2 ) predicted protein molecular weight of 29.8kDa.
[0063] (2) Purify the above protein using a nickel column:
[0064] 1. Equilibrate the column with buffer solution A (50mM Tris / HCl, pH 7.9, 0.5M NaCl) at a flow rate of 1mL / min.
[0065] 2. Load the sample at a flow rate of 1 mL / min, and collect the breakthrough.
[0066] 3. Wash the column with buffer solution A, flow rate 1mL / min, wash 40mL,
[0067] 4. Elution with buffer solution A+20mM imidazole, flow rate 1mL / min, wash 40ml, collect one tube every 4min.
[0068] 5. Collect samples for G250 detection to see if there is any protein eluted in the last tube. If there is no protein, proceed to th...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com